Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans
- PMID: 18815310
- PMCID: PMC2583677
- DOI: 10.1128/JVI.01623-08
Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans
Abstract
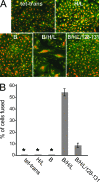
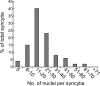
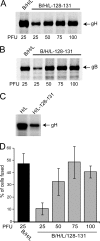
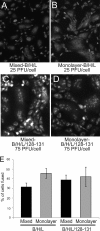
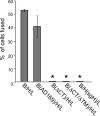
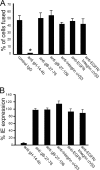
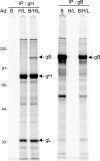
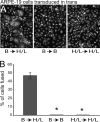
Herpesviruses use a cascade of interactions with different cell surface molecules to gain entry into cells. In many cases, this involves binding to abundant glycosaminoglycans or integrins followed by interactions with more limited cell surface proteins, leading to fusion with cellular membranes. Human cytomegalovirus (HCMV) has the ability to infect a wide variety of human cell types in vivo. However, very little is known about which HCMV glycoproteins mediate entry into various cell types, including relevant epithelial and endothelial cells. For other herpesviruses, studies of cell-cell fusion induced by viral proteins have provided substantial information about late stages of entry. In this report, we describe the fusion of epithelial, endothelial, microglial, and fibroblast cells in which HCMV gB and gH/gL were expressed from nonreplicating adenovirus vectors. Fusion frequently involved the majority of cells, and gB and gH/gL were both necessary and sufficient for fusion, whereas no fusion occurred when either glycoprotein was omitted. Coexpression of UL128, UL130, and UL131 did not enhance fusion. We concluded that the HCMV core fusion machinery consists of gB and gH/gL. Coimmunoprecipitation indicated that HCMV gB and gH/gL can interact. Importantly, expression of gB and gH/gL in trans (gB-expressing cells mixed with other gH/gL-expressing cells) resulted in substantial fusion. We believe that this is the first description of a multicomponent viral fusion machine that can be split between cells. If gB and gH/gL must interact for fusion, then these molecules must reach across the space between apposing cells. Expression of gB and gH/gL in trans with different cell types revealed surface molecules that are required for fusion on HCMV-permissive cells but not on nonpermissive cells.
Figures
Similar articles
-
Human Cytomegalovirus gH/gL/gO Promotes the Fusion Step of Entry into All Cell Types, whereas gH/gL/UL128-131 Broadens Virus Tropism through a Distinct Mechanism.J Virol. 2015 Sep;89(17):8999-9009. doi: 10.1128/JVI.01325-15. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085146 Free PMC article.
-
Scanning Mutagenesis of Human Cytomegalovirus Glycoprotein gH/gL.J Virol. 2015 Dec 9;90(5):2294-305. doi: 10.1128/JVI.01875-15. J Virol. 2015. PMID: 26656708 Free PMC article.
-
Human cytomegalovirus (HCMV) glycoprotein gB promotes virus entry in trans acting as the viral fusion protein rather than as a receptor-binding protein.mBio. 2013 Jun 4;4(3):e00332-13. doi: 10.1128/mBio.00332-13. mBio. 2013. PMID: 23736286 Free PMC article.
-
Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications.Rev Med Virol. 2010 May;20(3):136-55. doi: 10.1002/rmv.645. Rev Med Virol. 2010. PMID: 20084641 Review.
-
Pathogen at the Gates: Human Cytomegalovirus Entry and Cell Tropism.Viruses. 2018 Dec 11;10(12):704. doi: 10.3390/v10120704. Viruses. 2018. PMID: 30544948 Free PMC article. Review.
Cited by
-
Human Cytomegalovirus Envelope Protein gpUL132 Regulates Infectious Virus Production through Formation of the Viral Assembly Compartment.mBio. 2020 Sep 29;11(5):e02044-20. doi: 10.1128/mBio.02044-20. mBio. 2020. PMID: 32994323 Free PMC article.
-
Regulation of herpes simplex virus gB-induced cell-cell fusion by mutant forms of gH/gL in the absence of gD and cellular receptors.mBio. 2013 Feb 26;4(2):e00046-13. doi: 10.1128/mBio.00046-13. mBio. 2013. PMID: 23443004 Free PMC article.
-
Guinea pig cytomegalovirus trimer complex gH/gL/gO uses PDGFRA as universal receptor for cell fusion and entry.Virology. 2020 Sep;548:236-249. doi: 10.1016/j.virol.2020.05.012. Epub 2020 Jun 11. Virology. 2020. PMID: 32791352 Free PMC article.
-
Comparative analysis of gO isoforms reveals that strains of human cytomegalovirus differ in the ratio of gH/gL/gO and gH/gL/UL128-131 in the virion envelope.J Virol. 2013 Sep;87(17):9680-90. doi: 10.1128/JVI.01167-13. Epub 2013 Jun 26. J Virol. 2013. PMID: 23804643 Free PMC article.
-
Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB.J Virol. 2010 Dec;84(23):12292-9. doi: 10.1128/JVI.01700-10. Epub 2010 Sep 22. J Virol. 2010. PMID: 20861251 Free PMC article.
References
-
- Adler, B., L. Scrivano, Z. Ruzcics, B. Rupp, C. Sinzger, and U. Koszinowski. 2006. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J. Gen. Virol. 872451-2460. - PubMed
-
- Alford, C. A., and W. J. Britt. 1990. Cytomegalovirus, p. 1981-2010. In B. N. Fields and D. M. Knipe (ed.), Fields virology. Raven Press Ltd., New York, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

