Molecular dynamics simulation and coarse-grained analysis of the Arp2/3 complex
- PMID: 18805923
- PMCID: PMC2586551
- DOI: 10.1529/biophysj.108.143313
Molecular dynamics simulation and coarse-grained analysis of the Arp2/3 complex
Abstract
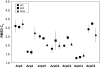
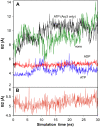
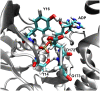
A molecular dynamics investigation and coarse-grained analysis of inactivated actin-related protein (Arp) 2/3 complex is presented. It was found that the nucleotide binding site within Arp3 remained in a closed position with bound ATP or ADP, but opened when simulation with no nucleotide was performed. In contrast, simulation of the isolated Arp3 subunit with bound ATP, showed a fast opening of the nucleotide binding cleft. A homology model for the missing subdomains 1 and 2 of Arp2 was constructed, and it was also found that the Arp2 binding cleft remained closed with bound nucleotide. Within the nucleotide binding cleft a distinct opening and closing period of 10 ns was observed in many of the simulations of Arp2/3 as well as isolated Arp3. Substitution studies were employed, and several alanine substitutions were found to induce a partial opening of the ATP binding cleft in Arp3 and Arp2, whereas only a single substitution was found to induce opening of the ADP binding cleft. It was also found that the nucleotide type did not cause a substantial change on interfacial contacts between Arp3 and the ArpC2, ArpC3 and ArpC4 subunits. Nucleotide-free Arp3 had generally less stable contacts, but the overall contact architecture was constant. Finally, nucleotide-dependent coarse-grained models for Arp3 are developed that serve to further highlight the structural differences induced in Arp3 by nucleotide hydrolysis.
Figures
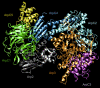
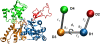


Similar articles
-
Nucleotide- and activator-dependent structural and dynamic changes of arp2/3 complex monitored by hydrogen/deuterium exchange and mass spectrometry.J Mol Biol. 2009 Jul 17;390(3):414-27. doi: 10.1016/j.jmb.2009.03.028. Epub 2009 Mar 17. J Mol Biol. 2009. PMID: 19298826 Free PMC article.
-
Nucleotide-mediated conformational changes of monomeric actin and Arp3 studied by molecular dynamics simulations.J Mol Biol. 2008 Feb 8;376(1):166-83. doi: 10.1016/j.jmb.2007.11.068. Epub 2007 Nov 28. J Mol Biol. 2008. PMID: 18155236 Free PMC article.
-
Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex.Mol Cell. 2007 May 11;26(3):449-57. doi: 10.1016/j.molcel.2007.04.017. Mol Cell. 2007. PMID: 17499050 Free PMC article.
-
Visualizing Arp2/3 complex activation mediated by binding of ATP and WASp using structural mass spectrometry.Proc Natl Acad Sci U S A. 2007 Jan 30;104(5):1552-7. doi: 10.1073/pnas.0605380104. Epub 2007 Jan 24. Proc Natl Acad Sci U S A. 2007. PMID: 17251352 Free PMC article.
-
Conformational changes in Arp2/3 complex induced by ATP, WASp-VCA, and actin filaments.Proc Natl Acad Sci U S A. 2018 Sep 11;115(37):E8642-E8651. doi: 10.1073/pnas.1717594115. Epub 2018 Aug 27. Proc Natl Acad Sci U S A. 2018. PMID: 30150414 Free PMC article.
Cited by
-
Structure and dynamics of the actin filament.J Mol Biol. 2010 Feb 19;396(2):252-63. doi: 10.1016/j.jmb.2009.11.034. Epub 2009 Nov 18. J Mol Biol. 2010. PMID: 19931282 Free PMC article.
-
Nucleotide- and activator-dependent structural and dynamic changes of arp2/3 complex monitored by hydrogen/deuterium exchange and mass spectrometry.J Mol Biol. 2009 Jul 17;390(3):414-27. doi: 10.1016/j.jmb.2009.03.028. Epub 2009 Mar 17. J Mol Biol. 2009. PMID: 19298826 Free PMC article.
-
Structural Basis of Arp2/3 Complex Inhibition by GMF, Coronin, and Arpin.J Mol Biol. 2017 Jan 20;429(2):237-248. doi: 10.1016/j.jmb.2016.11.030. Epub 2016 Dec 6. J Mol Biol. 2017. PMID: 27939292 Free PMC article.
-
Phosphorylation of the Arp2 subunit relieves auto-inhibitory interactions for Arp2/3 complex activation.PLoS Comput Biol. 2011 Nov;7(11):e1002226. doi: 10.1371/journal.pcbi.1002226. Epub 2011 Nov 10. PLoS Comput Biol. 2011. PMID: 22125478 Free PMC article.
-
Key structural features of the actin filament Arp2/3 complex branch junction revealed by molecular simulation.J Mol Biol. 2012 Feb 10;416(1):148-61. doi: 10.1016/j.jmb.2011.12.025. Epub 2011 Dec 17. J Mol Biol. 2012. PMID: 22206989 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

