Measles viruses possessing the polymerase protein genes of the Edmonston vaccine strain exhibit attenuated gene expression and growth in cultured cells and SLAM knock-in mice
- PMID: 18799577
- PMCID: PMC2583675
- DOI: 10.1128/JVI.00867-08
Measles viruses possessing the polymerase protein genes of the Edmonston vaccine strain exhibit attenuated gene expression and growth in cultured cells and SLAM knock-in mice
Abstract
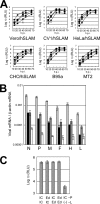
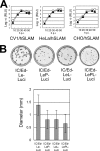
Live attenuated vaccines against measles have been developed through adaptation of clinical isolates of measles virus (MV) in various cultured cells. Analyses using recombinant MVs with chimeric genomes between wild-type and Edmonston vaccine strains indicated that viruses possessing the polymerase protein genes of the Edmonston strain exhibited attenuated viral gene expression and growth in cultured cells as well as in mice expressing an MV receptor, signaling lymphocyte activation molecule, regardless of whether the virus genome had the wild-type or vaccine-type promoter sequence. These data demonstrate that the polymerase protein genes of the Edmonston strain contribute to its attenuated phenotype.
Figures

Similar articles
-
Altered interaction of the matrix protein with the cytoplasmic tail of hemagglutinin modulates measles virus growth by affecting virus assembly and cell-cell fusion.J Virol. 2007 Jul;81(13):6827-36. doi: 10.1128/JVI.00248-07. Epub 2007 Apr 18. J Virol. 2007. PMID: 17442724 Free PMC article.
-
A molecularly cloned Schwarz strain of measles virus vaccine induces strong immune responses in macaques and transgenic mice.J Virol. 2003 Nov;77(21):11546-54. doi: 10.1128/jvi.77.21.11546-11554.2003. J Virol. 2003. PMID: 14557640 Free PMC article.
-
SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein.J Virol. 2002 Jul;76(13):6743-9. doi: 10.1128/jvi.76.13.6743-6749.2002. J Virol. 2002. PMID: 12050387 Free PMC article.
-
Measles virus receptors: SLAM and CD46.Rev Med Virol. 2004 Jul-Aug;14(4):217-29. doi: 10.1002/rmv.430. Rev Med Virol. 2004. PMID: 15248250 Review.
-
Measles virus.Hum Vaccin Immunother. 2015;11(1):21-6. doi: 10.4161/hv.34298. Epub 2014 Nov 1. Hum Vaccin Immunother. 2015. PMID: 25483511 Free PMC article. Review.
Cited by
-
What's going on with measles?J Virol. 2024 Aug 20;98(8):e0075824. doi: 10.1128/jvi.00758-24. Epub 2024 Jul 23. J Virol. 2024. PMID: 39041786 Review.
-
Infection of Pro- and Anti-Inflammatory Macrophages by Wild Type and Vaccine Strains of Measles Virus: NLRP3 Inflammasome Activation Independent of Virus Production.Viruses. 2023 Jan 17;15(2):260. doi: 10.3390/v15020260. Viruses. 2023. PMID: 36851476 Free PMC article.
-
Serotypic evolution of measles virus is constrained by multiple co-dominant B cell epitopes on its surface glycoproteins.Cell Rep Med. 2021 Mar 30;2(4):100225. doi: 10.1016/j.xcrm.2021.100225. eCollection 2021 Apr 20. Cell Rep Med. 2021. PMID: 33948566 Free PMC article.
-
Humanized Mice for Live-Attenuated Vaccine Research: From Unmet Potential to New Promises.Vaccines (Basel). 2020 Jan 21;8(1):36. doi: 10.3390/vaccines8010036. Vaccines (Basel). 2020. PMID: 31973073 Free PMC article. Review.
-
Evolution of Attenuation and Risk of Reversal in Peste des Petits Ruminants Vaccine Strain Nigeria 75/1.Viruses. 2019 Aug 7;11(8):724. doi: 10.3390/v11080724. Viruses. 2019. PMID: 31394790 Free PMC article.
References
-
- Bankamp, B., G. Hodge, M. B. McChesney, W. J. Bellini, and P. A. Rota. 2008. Genetic changes that affect the virulence of measles virus in a rhesus macaque model. Virology 37339-50. - PubMed
-
- Bankamp, B., J. Wilson, W. J. Bellini, and P. A. Rota. 2005. Identification of naturally occurring amino acid variations that affect the ability of the measles virus C protein to regulate genome replication and transcription. Virology 336120-129. - PubMed
-
- Borges, M. B., E. Caride, A. V. Jabor, J. M. Malachias, M. S. Freire, A. Homma, and R. Galler. 2008. Study of the genetic stability of measles virus CAM-70 vaccine strain after serial passages in chicken embryo fibroblasts primary cultures. Virus Genes 3635-44. - PubMed
-
- Caignard, G., M. Guerbois, J. L. Labernardiere, Y. Jacob, L. M. Jones, F. Wild, F. Tangy, and P. O. Vidalain. 2007. Measles virus V protein blocks Jak1-mediated phosphorylation of STAT1 to escape IFN-alpha/beta signaling. Virology 368351-362. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

