Impaired survival of peripheral T cells, disrupted NK/NKT cell development, and liver failure in mice lacking Gimap5
- PMID: 18796632
- PMCID: PMC2597598
- DOI: 10.1182/blood-2008-03-146555
Impaired survival of peripheral T cells, disrupted NK/NKT cell development, and liver failure in mice lacking Gimap5
Abstract
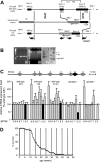
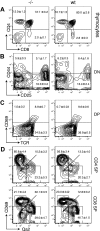
The loss of Gimap5 (GTPase of the immune-associated protein 5) gene function is the underlying cause of lymphopenia and autoimmune diabetes in the BioBreeding (BB) rat. The in vivo function of murine gimap5 is largely unknown. We show that selective gene ablation of the mouse gimap5 gene impairs the final intrathymic maturation of CD8 and CD4 T cells and compromises the survival of postthymic CD4 and CD8 cells, replicating findings in the BB rat model. In addition, gimap5 deficiency imposes a block of natural killer (NK)- and NKT-cell differentiation. Development of NK/NKT cells is restored on transfer of gimap5(-/-) bone marrow into a wild-type environment. Mice lacking gimap5 have a median survival of 15 weeks, exhibit chronic hepatic hematopoiesis, and in later stages show pronounced hepatocyte apoptosis, leading to liver failure. This pathology persists in a Rag2-deficient background in the absence of mature B, T, or NK cells and cannot be adoptively transferred by transplanting gimap5(-/-) bone marrow into wild-type recipients. We conclude that mouse gimap5 is necessary for the survival of peripheral T cells, NK/NKT-cell development, and the maintenance of normal liver function. These functions involve cell-intrinsic as well as cell-extrinsic mechanisms.
Figures




Similar articles
-
Critical role for Gimap5 in the survival of mouse hematopoietic stem and progenitor cells.J Exp Med. 2011 May 9;208(5):923-35. doi: 10.1084/jem.20101192. Epub 2011 Apr 18. J Exp Med. 2011. PMID: 21502331 Free PMC article.
-
CHOP mediates endoplasmic reticulum stress-induced apoptosis in Gimap5-deficient T cells.PLoS One. 2009;4(5):e5468. doi: 10.1371/journal.pone.0005468. Epub 2009 May 8. PLoS One. 2009. PMID: 19424493 Free PMC article.
-
Loss of T cell and B cell quiescence precedes the onset of microbial flora-dependent wasting disease and intestinal inflammation in Gimap5-deficient mice.J Immunol. 2010 Apr 1;184(7):3743-54. doi: 10.4049/jimmunol.0903164. Epub 2010 Feb 26. J Immunol. 2010. PMID: 20190135 Free PMC article.
-
Central role of gimap5 in maintaining peripheral tolerance and T cell homeostasis in the gut.Mediators Inflamm. 2015;2015:436017. doi: 10.1155/2015/436017. Epub 2015 Apr 7. Mediators Inflamm. 2015. PMID: 25944983 Free PMC article. Review.
-
The GIMAP Family Proteins: An Incomplete Puzzle.Front Immunol. 2021 May 31;12:679739. doi: 10.3389/fimmu.2021.679739. eCollection 2021. Front Immunol. 2021. PMID: 34135906 Free PMC article. Review.
Cited by
-
Whole genome sequencing reveals signals of adaptive admixture in Creole cattle.Sci Rep. 2023 Jul 27;13(1):12155. doi: 10.1038/s41598-023-38774-7. Sci Rep. 2023. PMID: 37500674 Free PMC article.
-
Changes in immune cell frequencies in primary and secondary lymphatic organs of LEW.1AR1-iddm rats, a model of human type 1 diabetes compared to other MHC congenic LEW inbred strains.Immunol Res. 2018 Aug;66(4):462-470. doi: 10.1007/s12026-018-9015-6. Immunol Res. 2018. PMID: 30143971
-
Critical role for Gimap5 in the survival of mouse hematopoietic stem and progenitor cells.J Exp Med. 2011 May 9;208(5):923-35. doi: 10.1084/jem.20101192. Epub 2011 Apr 18. J Exp Med. 2011. PMID: 21502331 Free PMC article.
-
Gimap5-dependent inactivation of GSK3β is required for CD4+ T cell homeostasis and prevention of immune pathology.Nat Commun. 2018 Jan 30;9(1):430. doi: 10.1038/s41467-018-02897-7. Nat Commun. 2018. PMID: 29382851 Free PMC article.
-
CHOP mediates endoplasmic reticulum stress-induced apoptosis in Gimap5-deficient T cells.PLoS One. 2009;4(5):e5468. doi: 10.1371/journal.pone.0005468. Epub 2009 May 8. PLoS One. 2009. PMID: 19424493 Free PMC article.
References
-
- Stamm O, Krucken J, Schmitt-Wrede HP, Benten WP, Wunderlich F. Human ortholog to mouse gene imap38 encoding an ER-localizable G-protein belongs to a gene family clustered on chromosome 7q32-36. Gene. 2002;282:159–167. - PubMed
-
- Andersen UN, Markholst H, Hornum L. The antiapoptotic gene Ian4l1 in the rat: genomic organization and promoter characterization. Gene. 2004;341:141–148. - PubMed
-
- Payne F, Smyth DJ, Pask R, et al. Haplotype tag single nucleotide polymorphism analysis of the human orthologues of the rat type 1 diabetes genes Ian4 (Lyp/Iddm1) and Cblb. Diabetes. 2004;53:505–509. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AI042380-100006/AI/NIAID NIH HHS/United States
- R01EB001421/EB/NIBIB NIH HHS/United States
- R01 AI073731/AI/NIAID NIH HHS/United States
- R01 EB001421/EB/NIBIB NIH HHS/United States
- AI42380-04/AI/NIAID NIH HHS/United States
- P01 AI042380/AI/NIAID NIH HHS/United States
- P01 AI042380-130006/AI/NIAID NIH HHS/United States
- R01 HL073284/HL/NHLBI NIH HHS/United States
- P01 AI042380-110006/AI/NIAID NIH HHS/United States
- P01 AI042380-120006/AI/NIAID NIH HHS/United States
- HL073284/HL/NHLBI NIH HHS/United States
- R01A1064826-01/PHS HHS/United States
- R01 AI047154/AI/NIAID NIH HHS/United States
- R01AI47154/AI/NIAID NIH HHS/United States
- P01 AI042380-090006/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

