Transforming growth factor beta-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth
- PMID: 18794361
- PMCID: PMC2573298
- DOI: 10.1128/MCB.01192-08
Transforming growth factor beta-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth
Abstract
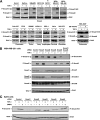
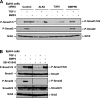
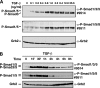
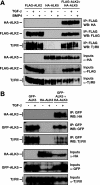
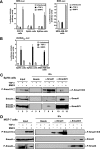
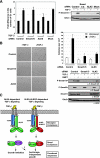
Transforming growth factor beta (TGF-beta) signals predominantly through a receptor complex comprising ALK5 and TbetaRII to activate receptor-regulated Smads (R-Smads) Smad2 and Smad3. In endothelial cells, however, TGF-beta can additionally activate Smad1 and Smad5. Here, we report that TGF-beta also strongly induces phosphorylation of Smad1/5 in many different normal epithelial cells, epithelium-derived tumor cells, and fibroblasts. We demonstrate that TbetaRII and ALK5, as well as ALK2 and/or ALK3, are required for TGF-beta-induced Smad1/5 phosphorylation. We show that the simultaneous activation of the R-Smads Smad2/3 and Smad1/5 by TGF-beta results in the formation of mixed R-Smad complexes, containing, for example, phosphorylated Smad1 and Smad2. The prevalence of these mixed R-Smad complexes explains why TGF-beta-induced Smad1/5 phosphorylation does not result in transcriptional activation via bone morphogenetic protein (BMP)-responsive elements, which bind activated Smad1/5-Smad4 complexes that are induced by BMP stimulation. Thus, TGF-beta induces two parallel pathways: one signaling via Smad2-Smad4 or Smad3-Smad4 complexes and the other signaling via mixed R-Smad complexes. Finally, we assess the function of the novel arm of TGF-beta signaling and show that TGF-beta-induced Smad1/5 activation is not required for the growth-inhibitory effects of TGF-beta but is specifically required for TGF-beta-induced anchorage-independent growth.
Figures

Similar articles
-
Transforming growth factor β inhibits bone morphogenetic protein-induced transcription through novel phosphorylated Smad1/5-Smad3 complexes.Mol Cell Biol. 2012 Jul;32(14):2904-16. doi: 10.1128/MCB.00231-12. Epub 2012 May 21. Mol Cell Biol. 2012. PMID: 22615489 Free PMC article.
-
Transforming growth factor-β stimulates Smad1/5 signaling in pulmonary artery smooth muscle cells and fibroblasts of the newborn mouse through ALK1.Am J Physiol Lung Cell Mol Physiol. 2017 Sep 1;313(3):L615-L627. doi: 10.1152/ajplung.00079.2017. Epub 2017 Jun 22. Am J Physiol Lung Cell Mol Physiol. 2017. PMID: 28642261 Free PMC article.
-
Growth differentiation factor-9 signaling is mediated by the type I receptor, activin receptor-like kinase 5.Mol Endocrinol. 2004 Mar;18(3):653-65. doi: 10.1210/me.2003-0393. Epub 2003 Dec 18. Mol Endocrinol. 2004. PMID: 14684852
-
Smad regulation in TGF-beta signal transduction.J Cell Sci. 2001 Dec;114(Pt 24):4359-69. doi: 10.1242/jcs.114.24.4359. J Cell Sci. 2001. PMID: 11792802 Review.
-
Transforming growth factor-β signalling: role and consequences of Smad linker region phosphorylation.Cell Signal. 2013 Oct;25(10):2017-24. doi: 10.1016/j.cellsig.2013.06.001. Epub 2013 Jun 11. Cell Signal. 2013. PMID: 23770288 Review.
Cited by
-
Beyond TGFβ: roles of other TGFβ superfamily members in cancer.Nat Rev Cancer. 2013 May;13(5):328-41. doi: 10.1038/nrc3500. Nat Rev Cancer. 2013. PMID: 23612460 Free PMC article. Review.
-
Morphogenesis and cell fate determination within the adaxial cell equivalence group of the zebrafish myotome.PLoS Genet. 2012;8(10):e1003014. doi: 10.1371/journal.pgen.1003014. Epub 2012 Oct 25. PLoS Genet. 2012. PMID: 23133395 Free PMC article.
-
Immune-mediated pathology in Duchenne muscular dystrophy.Sci Transl Med. 2015 Aug 5;7(299):299rv4. doi: 10.1126/scitranslmed.aaa7322. Sci Transl Med. 2015. PMID: 26246170 Free PMC article. Review.
-
Computational modelling of Smad-mediated negative feedback and crosstalk in the TGF-β superfamily network.J R Soc Interface. 2013 Jun 26;10(86):20130363. doi: 10.1098/rsif.2013.0363. Print 2013 Sep 6. J R Soc Interface. 2013. PMID: 23804438 Free PMC article.
-
Engineering of a Microscale Niche for Pancreatic Tumor Cells Using Bioactive Film Coatings Combined with 3D-Architectured Scaffolds.ACS Appl Mater Interfaces. 2022 Mar 23;14(11):13107-13121. doi: 10.1021/acsami.2c01747. Epub 2022 Mar 11. ACS Appl Mater Interfaces. 2022. PMID: 35275488 Free PMC article.
References
-
- Bardwell, V. J., and R. Treisman. 1994. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 81664-1677. - PubMed
-
- Byfield, S. D., and A. B. Roberts. 2004. Lateral signaling enhances TGF-β response complexity. Trends Cell Biol. 14107-111. - PubMed
-
- Chen, X., M. J. Rubock, and M. Whitman. 1996. A transcriptional partner for MAD proteins in TGF-β signalling. Nature 383691-696. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
