Dicistronic tRNA-5S rRNA genes in Yarrowia lipolytica: an alternative TFIIIA-independent way for expression of 5S rRNA genes
- PMID: 18790808
- PMCID: PMC2566860
- DOI: 10.1093/nar/gkn549
Dicistronic tRNA-5S rRNA genes in Yarrowia lipolytica: an alternative TFIIIA-independent way for expression of 5S rRNA genes
Abstract
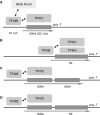
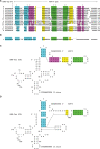
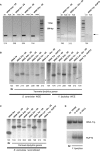
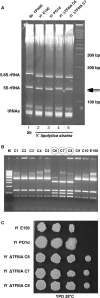
In eukaryotes, genes transcribed by RNA polymerase III (Pol III) carry their own internal promoters and as such, are transcribed as individual units. Indeed, a very few cases of dicistronic Pol III genes are yet known. In contrast to other hemiascomycetes, 5S rRNA genes of Yarrowia lipolytica are not embedded into the tandemly repeated rDNA units, but appear scattered throughout the genome. We report here an unprecedented genomic organization: 48 over the 108 copies of the 5S rRNA genes are located 3' of tRNA genes. We show that these peculiar tRNA-5S rRNA dicistronic genes are expressed in vitro and in vivo as Pol III transcriptional fusions without the need of the 5S rRNA gene-specific factor TFIIIA, the deletion of which displays a viable phenotype. We also report the existence of a novel putative non-coding Pol III RNA of unknown function about 70 nucleotide-long (RUF70), the 13 genes of which are devoid of internal Pol III promoters and located 3' of the 13 copies of the tDNA-Trp (CCA). All genes embedded in the various dicistronic genes, fused 5S rRNA genes, RUF70 genes and their leader tRNA genes appear to be efficiently transcribed and their products correctly processed in vivo.
Figures

Similar articles
-
The Dictyostelium discoideum 5S rDNA is organized in the same transcriptional orientation as the other rDNAs.Biochem Biophys Res Commun. 1993 Mar 15;191(2):558-64. doi: 10.1006/bbrc.1993.1254. Biochem Biophys Res Commun. 1993. PMID: 8461013
-
The only essential function of TFIIIA in yeast is the transcription of 5S rRNA genes.Proc Natl Acad Sci U S A. 1995 Sep 26;92(20):9338-42. doi: 10.1073/pnas.92.20.9338. Proc Natl Acad Sci U S A. 1995. PMID: 7568129 Free PMC article.
-
Alternative splicing of anciently exonized 5S rRNA regulates plant transcription factor TFIIIA.Genome Res. 2009 May;19(5):913-21. doi: 10.1101/gr.086876.108. Epub 2009 Feb 10. Genome Res. 2009. PMID: 19211543 Free PMC article.
-
Structure, function and regulation of Transcription Factor IIIA: From Xenopus to Arabidopsis.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):274-82. doi: 10.1016/j.bbagrm.2012.10.013. Epub 2012 Nov 8. Biochim Biophys Acta. 2013. PMID: 23142779 Review.
-
Ribosomal 5S RNA from Xenopus laevis oocytes: conformation and interaction with transcription factor IIIA.Biochimie. 1990 Jun-Jul;72(6-7):437-52. doi: 10.1016/0300-9084(90)90068-r. Biochimie. 1990. PMID: 2124147 Review.
Cited by
-
Genetic characterization of Plectorhinchus mediterraneus yields important clues about genome organization and evolution of multigene families.BMC Genet. 2012 Apr 30;13:33. doi: 10.1186/1471-2156-13-33. BMC Genet. 2012. PMID: 22545758 Free PMC article.
-
Detection and analysis of alternative splicing in Yarrowia lipolytica reveal structural constraints facilitating nonsense-mediated decay of intron-retaining transcripts.Genome Biol. 2010;11(6):R65. doi: 10.1186/gb-2010-11-6-r65. Epub 2010 Jun 23. Genome Biol. 2010. PMID: 20573210 Free PMC article.
-
Widespread occurrence of non-canonical transcription termination by human RNA polymerase III.Nucleic Acids Res. 2011 Jul;39(13):5499-512. doi: 10.1093/nar/gkr074. Epub 2011 Mar 17. Nucleic Acids Res. 2011. PMID: 21421562 Free PMC article.
-
Evolutionary dynamics of the 5S rDNA gene family in the mussel Mytilus: mixed effects of birth-and-death and concerted evolution.J Mol Evol. 2010 May;70(5):413-26. doi: 10.1007/s00239-010-9341-3. Epub 2010 Apr 13. J Mol Evol. 2010. PMID: 20386892
-
Biotransformation of acetophenone and its halogen derivatives by Yarrowia lipolytica strains.Ann Microbiol. 2015;65(2):1097-1107. doi: 10.1007/s13213-014-0955-3. Epub 2014 Aug 22. Ann Microbiol. 2015. PMID: 26005401 Free PMC article.
References
-
- Blumenthal T. Operons in eukaryotes. Brief. Funct. Genomic. Proteomic. 2004;3:199–211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

