Phosphate coordination and movement of DNA in the Tn5 synaptic complex: role of the (R)YREK motif
- PMID: 18790806
- PMCID: PMC2566895
- DOI: 10.1093/nar/gkn577
Phosphate coordination and movement of DNA in the Tn5 synaptic complex: role of the (R)YREK motif
Abstract
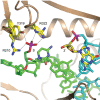
Bacterial DNA transposition is an important model system for studying DNA recombination events such as HIV-1 DNA integration and RAG-1-mediated V(D)J recombination. This communication focuses on the role of protein-phosphate contacts in manipulating DNA structure as a requirement for transposition catalysis. In particular, the participation of the nontransferred strand (NTS) 5' phosphate in Tn5 transposition strand transfer is analyzed. The 5' phosphate plays no direct catalytic role, nonetheless its presence stimulates strand transfer approximately 30-fold. X-ray crystallography indicates that transposase-DNA complexes formed with NTS 5' phosphorylated DNA have two properties that contrast with structures formed with complexes lacking the 5' phosphate or complexes generated from in-crystal hairpin cleavage. Transposase residues R210, Y319 and R322 of the (R)YREK motif coordinate the 5' phosphate rather than the subterminal NTS phosphate, and the 5' NTS end is moved away from the 3' transferred strand end. Mutation R210A impairs the 5' phosphate stimulation. It is posited that DNA phosphate coordination by R210, Y319 and R322 results in movement of the 5' NTS DNA away from the 3'-end thus allowing efficient target DNA binding. It is likely that this role for the newly identified RYR triad is utilized by other transposase-related proteins.
Figures

Similar articles
-
Three-dimensional structure of the Tn5 synaptic complex transposition intermediate.Science. 2000 Jul 7;289(5476):77-85. doi: 10.1126/science.289.5476.77. Science. 2000. PMID: 10884228
-
Tn5 transposase loops DNA in the absence of Tn5 transposon end sequences.Mol Microbiol. 2006 Dec;62(6):1558-68. doi: 10.1111/j.1365-2958.2006.05471.x. Mol Microbiol. 2006. PMID: 17074070
-
Mutation of Tn5 transposase beta-loop residues affects all steps of Tn5 transposition: the role of conformational changes in Tn5 transposition.Biochemistry. 2006 Dec 26;45(51):15552-62. doi: 10.1021/bi061227v. Epub 2006 Dec 5. Biochemistry. 2006. PMID: 17176076 Free PMC article.
-
Structure/function insights into Tn5 transposition.Curr Opin Struct Biol. 2004 Feb;14(1):50-7. doi: 10.1016/j.sbi.2004.01.008. Curr Opin Struct Biol. 2004. PMID: 15102449 Review.
-
Tn5 as a model for understanding DNA transposition.Mol Microbiol. 2003 Mar;47(5):1199-206. doi: 10.1046/j.1365-2958.2003.03382.x. Mol Microbiol. 2003. PMID: 12603728 Review.
Cited by
-
DNA Transposition at Work.Chem Rev. 2016 Oct 26;116(20):12758-12784. doi: 10.1021/acs.chemrev.6b00003. Epub 2016 May 17. Chem Rev. 2016. PMID: 27187082 Free PMC article. Review.
-
Target capture during Mos1 transposition.J Biol Chem. 2014 Jan 3;289(1):100-11. doi: 10.1074/jbc.M113.523894. Epub 2013 Nov 22. J Biol Chem. 2014. PMID: 24269942 Free PMC article.
-
DDE transposases: Structural similarity and diversity.Adv Drug Deliv Rev. 2010 Sep 30;62(12):1187-95. doi: 10.1016/j.addr.2010.06.006. Epub 2010 Jul 6. Adv Drug Deliv Rev. 2010. PMID: 20615441 Free PMC article. Review.
-
Efficient second strand cleavage during Holliday junction resolution by RuvC requires both increased junction flexibility and an exposed 5' phosphate.PLoS One. 2009;4(4):e5347. doi: 10.1371/journal.pone.0005347. Epub 2009 Apr 28. PLoS One. 2009. PMID: 19399178 Free PMC article.
-
Acinetobacter insertion sequence ISAba11 belongs to a novel family that encodes transposases with a signature HHEK motif.Appl Environ Microbiol. 2012 Jan;78(2):471-80. doi: 10.1128/AEM.05663-11. Epub 2011 Nov 11. Appl Environ Microbiol. 2012. PMID: 22081580 Free PMC article.
References
-
- Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA. Washington, DC: ASM Press; 2002.
-
- Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: Similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. - PubMed
-
- Rice P, Mizuuchi K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell. 1995;82:209–220. - PubMed
-
- Davies DR, Braam LM, Reznikoff WS, Rayment I. The three-dimensional structure of a Tn5 transposase-related protein determined to 2.9 Å resolution. J. Biol. Chem. 1999;274:11904–11913. - PubMed
-
- Gellert M. V(D)J recombination. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA. ASM Press: Washington, DC; 2002. pp. 705–729.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

