Viral cystatin evolution and three-dimensional structure modelling: a case of directional selection acting on a viral protein involved in a host-parasitoid interaction
- PMID: 18783611
- PMCID: PMC2553070
- DOI: 10.1186/1741-7007-6-38
Viral cystatin evolution and three-dimensional structure modelling: a case of directional selection acting on a viral protein involved in a host-parasitoid interaction
Abstract
Background: In pathogens, certain genes encoding proteins that directly interact with host defences coevolve with their host and are subject to positive selection. In the lepidopteran host-wasp parasitoid system, one of the most original strategies developed by the wasps to defeat host defences is the injection of a symbiotic polydnavirus at the same time as the wasp eggs. The virus is essential for wasp parasitism success since viral gene expression alters the immune system and development of the host. As a wasp mutualist symbiont, the virus is expected to exhibit a reduction in genome complexity and evolve under wasp phyletic constraints. However, as a lepidopteran host pathogenic symbiont, the virus is likely undergoing strong selective pressures for the acquisition of new functions by gene acquisition or duplication. To understand the constraints imposed by this particular system on virus evolution, we studied a polydnavirus gene family encoding cyteine protease inhibitors of the cystatin superfamily.
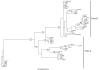
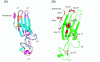
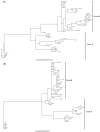
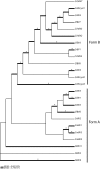
Results: We show that cystatins are the first bracovirus genes proven to be subject to strong positive selection within a host-parasitoid system. A generated three-dimensional model of Cotesia congregata bracovirus cystatin 1 provides a powerful framework to position positively selected residues and reveal that they are concentrated in the vicinity of actives sites which interact with cysteine proteases directly. In addition, phylogenetic analyses reveal two different cystatin forms which evolved under different selective constraints and are characterized by independent adaptive duplication events.
Conclusion: Positive selection acts to maintain cystatin gene duplications and induces directional divergence presumably to ensure the presence of efficient and adapted cystatin forms. Directional selection has acted on key cystatin active sites, suggesting that cystatins coevolve with their host target. We can strongly suggest that cystatins constitute major virulence factors, as was already proposed in previous functional studies.
Figures
Similar articles
-
When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses.Philos Trans R Soc Lond B Biol Sci. 2013 Aug 12;368(1626):20130051. doi: 10.1098/rstb.2013.0051. Print 2013 Sep 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23938758 Free PMC article. Review.
-
A virus essential for insect host-parasite interactions encodes cystatins.J Virol. 2005 Aug;79(15):9765-76. doi: 10.1128/JVI.79.15.9765-9776.2005. J Virol. 2005. PMID: 16014938 Free PMC article.
-
Identification of parasite-responsive cysteine proteases in Manduca sexta.Biol Chem. 2009 May-Jun;390(5-6):493-502. doi: 10.1515/BC.2009.061. Biol Chem. 2009. PMID: 19361282
-
Functional annotation of Cotesia congregata bracovirus: identification of viral genes expressed in parasitized host immune tissues.J Virol. 2014 Aug;88(16):8795-812. doi: 10.1128/JVI.00209-14. Epub 2014 May 28. J Virol. 2014. PMID: 24872581 Free PMC article.
-
Unfolding the evolutionary story of polydnaviruses.Virus Res. 2006 Apr;117(1):81-9. doi: 10.1016/j.virusres.2006.01.001. Epub 2006 Feb 3. Virus Res. 2006. PMID: 16460826 Review.
Cited by
-
When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses.Philos Trans R Soc Lond B Biol Sci. 2013 Aug 12;368(1626):20130051. doi: 10.1098/rstb.2013.0051. Print 2013 Sep 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23938758 Free PMC article. Review.
-
Recurrent Domestication by Lepidoptera of Genes from Their Parasites Mediated by Bracoviruses.PLoS Genet. 2015 Sep 17;11(9):e1005470. doi: 10.1371/journal.pgen.1005470. eCollection 2015 Sep. PLoS Genet. 2015. PMID: 26379286 Free PMC article.
-
Microbial inhibitors of cysteine proteases.Med Microbiol Immunol. 2016 Aug;205(4):275-96. doi: 10.1007/s00430-016-0454-1. Epub 2016 Apr 5. Med Microbiol Immunol. 2016. PMID: 27048482 Review.
-
Phylogenomic analysis of the cystatin superfamily in eukaryotes and prokaryotes.BMC Evol Biol. 2009 Nov 18;9:266. doi: 10.1186/1471-2148-9-266. BMC Evol Biol. 2009. PMID: 19919722 Free PMC article.
-
Polydnaviruses of Parasitic Wasps: Domestication of Viruses To Act as Gene Delivery Vectors.Insects. 2012 Jan 31;3(1):91-119. doi: 10.3390/insects3010091. Insects. 2012. PMID: 26467950 Free PMC article. Review.
References
-
- Dodds P, Lawrence GJ, Catanzariti A-M, Teh T, Wang C-IA, Ayliffe MA, Ellis J. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

