Human-specific gain of function in a developmental enhancer
- PMID: 18772437
- PMCID: PMC2658639
- DOI: 10.1126/science.1159974
Human-specific gain of function in a developmental enhancer
Abstract
Changes in gene regulation are thought to have contributed to the evolution of human development. However, in vivo evidence for uniquely human developmental regulatory function has remained elusive. In transgenic mice, a conserved noncoding sequence (HACNS1) that evolved extremely rapidly in humans acted as an enhancer of gene expression that has gained a strong limb expression domain relative to the orthologous elements from chimpanzee and rhesus macaque. This gain of function was consistent across two developmental stages in the mouse and included the presumptive anterior wrist and proximal thumb. In vivo analyses with synthetic enhancers, in which human-specific substitutions were introduced into the chimpanzee enhancer sequence or reverted in the human enhancer to the ancestral state, indicated that 13 substitutions clustered in an 81-base pair module otherwise highly constrained among terrestrial vertebrates were sufficient to confer the human-specific limb expression domain.
Figures


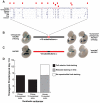
Comment in
-
Genetics. Enhancing gene regulation.Science. 2008 Sep 5;321(5894):1300-1. doi: 10.1126/science.1163568. Science. 2008. PMID: 18772422 No abstract available.
-
Comment on "Human-specific gain of function in a developmental enhancer".Science. 2009 Feb 6;323(5915):714; author reply 714. doi: 10.1126/science.1165848. Science. 2009. PMID: 19197042
Similar articles
-
Many human accelerated regions are developmental enhancers.Philos Trans R Soc Lond B Biol Sci. 2013 Nov 11;368(1632):20130025. doi: 10.1098/rstb.2013.0025. Print 2013 Dec 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 24218637 Free PMC article.
-
Two Types of Etiological Mutation in the Limb-Specific Enhancer of Shh.G3 (Bethesda). 2017 Sep 7;7(9):2991-2998. doi: 10.1534/g3.117.044669. G3 (Bethesda). 2017. PMID: 28710291 Free PMC article.
-
Functional analysis of limb transcriptional enhancers in the mouse.Evol Dev. 2014 Jul-Aug;16(4):207-23. doi: 10.1111/ede.12084. Epub 2014 Jun 11. Evol Dev. 2014. PMID: 24920384 Free PMC article.
-
Globalisation reaches gene regulation: the case for vertebrate limb development.Curr Opin Genet Dev. 2005 Aug;15(4):403-9. doi: 10.1016/j.gde.2005.06.011. Curr Opin Genet Dev. 2005. PMID: 15979301 Review.
-
The temporal dynamics of vertebrate limb development, teratogenesis and evolution.Curr Opin Genet Dev. 2010 Aug;20(4):384-90. doi: 10.1016/j.gde.2010.04.014. Epub 2010 May 27. Curr Opin Genet Dev. 2010. PMID: 20537528 Review.
Cited by
-
A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics.Elife. 2015 Jun 23;4:e07103. doi: 10.7554/eLife.07103. Elife. 2015. PMID: 26102527 Free PMC article.
-
Evolutionary Innovations in Conserved Regulatory Elements Associate With Developmental Genes in Mammals.Mol Biol Evol. 2024 Oct 4;41(10):msae199. doi: 10.1093/molbev/msae199. Mol Biol Evol. 2024. PMID: 39302728 Free PMC article.
-
Sequence divergence and retrotransposon insertion underlie interspecific epigenetic differences in primates.Mol Biol Evol. 2022 Oct 11;39(10):msac208. doi: 10.1093/molbev/msac208. Online ahead of print. Mol Biol Evol. 2022. PMID: 36219870 Free PMC article.
-
A cell type-aware framework for nominating non-coding variants in Mendelian regulatory disorders.medRxiv [Preprint]. 2023 Dec 27:2023.12.22.23300468. doi: 10.1101/2023.12.22.23300468. medRxiv. 2023. Update in: Nat Commun. 2024 Sep 27;15(1):8268. doi: 10.1038/s41467-024-52463-7 PMID: 38234731 Free PMC article. Updated. Preprint.
-
Contrasts between adaptive coding and noncoding changes during human evolution.Proc Natl Acad Sci U S A. 2010 Apr 27;107(17):7853-7. doi: 10.1073/pnas.0911249107. Epub 2010 Apr 12. Proc Natl Acad Sci U S A. 2010. PMID: 20385805 Free PMC article.
References
-
- Trinkaus E. Evolution of human manipulation. In: Jones S, Martin R, Pilbeam D, editors. The Cambridge Encyclopedia of Human Evolution. Cambridge Univ. Press; New York: 1993. pp. 346–349.
-
- Carroll SB. Nature. 2003;422:849. - PubMed
-
- King MC, Wilson AC. Science. 1975;188:107. - PubMed
-
- Tournamille C, et al. Nat. Genet. 1995;10:224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

