Genetic diversity in the yellow head nidovirus complex
- PMID: 18768192
- PMCID: PMC7103379
- DOI: 10.1016/j.virol.2008.07.005
Genetic diversity in the yellow head nidovirus complex
Abstract
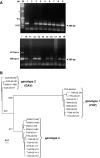
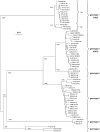
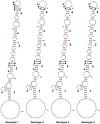
Penaeus monodon shrimp collected from across the Indo-Pacific region during 1997-2004 were screened for the presence of yellow head-related viruses. Phylogenetic analyses of amplified ORF1b gene segments identified at least six distinct genetic lineages (genotypes). Genotype 1 (YHV) was detected only in shrimp with yellow head disease. Genotype 2 (GAV) was detected in diseased shrimp with the less severe condition described as mid-crop mortality syndrome and in healthy shrimp from Australia, Thailand and Vietnam. Other genotypes occurred commonly in healthy shrimp. Sequence comparisons of structural protein genes (ORF2 and ORF3), intergenic regions (IGRs) and the long 3'-UTR supported the delineation of genotypes and identified both conserved and variant structural features. In putative transcription regulating sequences (TRSs) encompassing the sub-genomic mRNA 5'-termini, a core motif (5'-GUCAAUUACAAC-3') is absolutely conserved. A small (83 nt) open reading frame (ORF4) in the 3'-UTR of GAV is variously truncated in all other genotypes and a TRS-like element preceding ORF4 is invariably corrupted by a A>G/U substitution in the central core motif (5'-UU(G/U)CAAC-3'). The data support previous evidence that ORF4 is a non-functional gene under construction or deconstruction. The 3'-UTRs also contain predicted 3'-terminal hairpin-loop structures that are preserved in all genotypes by compensatory nucleotide substitutions, suggesting a role in polymerase recognition for minus-strand RNA synthesis.
Figures

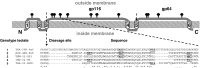
 ) on the illustration. Six predicted transmembrane-spanning domains (TM1–TM6) are numbered.
) on the illustration. Six predicted transmembrane-spanning domains (TM1–TM6) are numbered.
Similar articles
-
RNA transcription analysis and completion of the genome sequence of yellow head nidovirus.Virus Res. 2008 Sep;136(1-2):157-65. doi: 10.1016/j.virusres.2008.05.008. Epub 2008 Jun 11. Virus Res. 2008. PMID: 18582978 Free PMC article.
-
Complete genome sequence of an isolate of a novel genotype of yellow head virus from Fenneropenaeus chinensis indigenous in China.Arch Virol. 2017 Apr;162(4):1149-1152. doi: 10.1007/s00705-016-3203-2. Epub 2017 Jan 2. Arch Virol. 2017. PMID: 28044194
-
Homologous genetic recombination in the yellow head complex of nidoviruses infecting Penaeus monodon shrimp.Virology. 2009 Jul 20;390(1):79-88. doi: 10.1016/j.virol.2009.04.015. Epub 2009 May 31. Virology. 2009. PMID: 19487006 Free PMC article.
-
ICTV Virus Taxonomy Profile: Roniviridae.J Gen Virol. 2021 Jan;102(1):jgv001514. doi: 10.1099/jgv.0.001514. J Gen Virol. 2021. PMID: 33108263 Free PMC article. Review.
-
Rapid and sensitive detection of shrimp yellow head virus by loop-mediated isothermal amplification combined with a lateral flow dipstick.J Virol Methods. 2013 Mar;188(1-2):51-6. doi: 10.1016/j.jviromet.2012.11.041. Epub 2012 Dec 7. J Virol Methods. 2013. PMID: 23219929 Review.
Cited by
-
Evidence for a novel coding sequence overlapping the 5'-terminal approximately 90 codons of the gill-associated and yellow head okavirus envelope glycoprotein gene.Virol J. 2009 Dec 17;6:222. doi: 10.1186/1743-422X-6-222. Virol J. 2009. PMID: 20017924 Free PMC article.
-
The effects of Nigella sativa (Ns), Anthemis hyalina (Ah) and Citrus sinensis (Cs) extracts on the replication of coronavirus and the expression of TRP genes family.Mol Biol Rep. 2014 Mar;41(3):1703-11. doi: 10.1007/s11033-014-3019-7. Epub 2014 Jan 12. Mol Biol Rep. 2014. PMID: 24413991 Free PMC article.
-
Updated roles of the gut microbiota in exploring shrimp etiology, polymicrobial pathogens, and disease incidence.Zool Res. 2024 Jul 18;45(4):910-923. doi: 10.24272/j.issn.2095-8137.2024.158. Zool Res. 2024. PMID: 39021080 Free PMC article. Review.
-
Classification, replication, and transcription of Nidovirales.Front Microbiol. 2024 Jan 24;14:1291761. doi: 10.3389/fmicb.2023.1291761. eCollection 2023. Front Microbiol. 2024. PMID: 38328580 Free PMC article. Review.
-
Immunological-based assays for specific detection of shrimp viruses.World J Virol. 2014 Feb 12;3(1):1-10. doi: 10.5501/wjv.v3.i1.1. World J Virol. 2014. PMID: 24567913 Free PMC article. Review.
References
-
- Beerens N., Snijder E.J. RNA signals in the 3′-terminus of the genome of Equine arteritis virus are required for viral RNA synthesis. J. Gen. Virol. 2006;87:1977–1983. - PubMed
-
- Chantanachookin C., Boonyaratpalin S., Kasornchandra J., Direkbusaratana S., Ekpanithanpong U., Supamataya K., Sriurairatana S., Flegel T.W. Histology and untrastructure reveal a new granulosis-like virus in Penaeus monodon affected by yellow-head disease. Dis. Aquat. Org. 1993;17:145–157.
-
- Chayaburakul K., Nash G., Pratanpipat P., Sriurairatana S., Withyachumnarnkul B. Multiple pathogens found in growth-retarded black tiger shrimp Penaeus monodon cultivated in Thailand. Dis. Aquat. Org. 2004;60:89–96. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

