Similar membrane affinity of mono- and Di-S-acylated ras membrane anchors: a new twist in the role of protein lipidation
- PMID: 18761454
- PMCID: PMC2646725
- DOI: 10.1021/ja805110q
Similar membrane affinity of mono- and Di-S-acylated ras membrane anchors: a new twist in the role of protein lipidation
Abstract
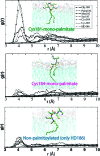
The functionally required membrane attachment of Ras is achieved through an invariant isoprenylation of a C-terminal Cys, supplemented by further lipid modification of adjacent Cys residues by one (N-ras) or two (H-ras) palmitoyls. However, whether the triply lipidated membrane anchor of H-ras has a higher membrane affinity than its doubly lipidated counterpart, or whether the affinity contribution of the two palmitates and the farnesyl is additive, was not known. To address this issue, we carried out potential of mean force (PMF or free energy profile) calculations on a hexadecylated but nonpalmitoylated anchor (Cys186-HD), hexadecylated and monopalmitoylated anchors (Cys181-monopalmitate and Cys184-monopalmitate), and a nonlipid-modified anchor. We found that the overall insertion free energy follows the trend Cys181/Cys184-bipalmitate (wild type) approximately Cys181-monopalmitate > Cys184-monopalmitate >> nonpalmitoylated anchor. Consistent with suggestions from recent cell biological experiments, the computed PMFs, coupled with structural analysis, demonstrate that membrane affinity of the Ras anchor depends on both the hydrophobicity of the palmitate and the prenyl groups and the spacing between them. The data further suggest that while Cys181-palmitate and Cys186-farnesyl together provide sufficient hydrophobic force for tight membrane binding, the palmitoyl at Cys184 is likely designed to serve another, presumably functional, role.
Figures
Similar articles
-
Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling.Mol Cell Biol. 2005 Aug;25(15):6722-33. doi: 10.1128/MCB.25.15.6722-6733.2005. Mol Cell Biol. 2005. PMID: 16024806 Free PMC article.
-
Insertion of lipidated Ras proteins into lipid monolayers studied by infrared reflection absorption spectroscopy (IRRAS).Biophys J. 2006 Aug 15;91(4):1388-401. doi: 10.1529/biophysj.106.084624. Epub 2006 May 26. Biophys J. 2006. PMID: 16731561 Free PMC article.
-
Influence of the lipid anchor motif of N-ras on the interaction with lipid membranes: a surface plasmon resonance study.Biophys J. 2010 May 19;98(10):2226-35. doi: 10.1016/j.bpj.2010.02.005. Biophys J. 2010. PMID: 20483331 Free PMC article.
-
Deciphering lipid codes: K-Ras as a paradigm.Traffic. 2018 Mar;19(3):157-165. doi: 10.1111/tra.12541. Epub 2017 Dec 10. Traffic. 2018. PMID: 29120102 Free PMC article. Review.
-
Membrane binding of lipidated Ras peptides and proteins--the structural point of view.Biochim Biophys Acta. 2009 Jan;1788(1):273-88. doi: 10.1016/j.bbamem.2008.08.006. Epub 2008 Aug 15. Biochim Biophys Acta. 2009. PMID: 18771652 Review.
Cited by
-
Organization, dynamics, and segregation of Ras nanoclusters in membrane domains.Proc Natl Acad Sci U S A. 2012 May 22;109(21):8097-102. doi: 10.1073/pnas.1200773109. Epub 2012 May 4. Proc Natl Acad Sci U S A. 2012. PMID: 22562795 Free PMC article.
-
Deformation of a Two-domain Lipid Bilayer due to Asymmetric Insertion of Lipid-modified Ras Peptides.Soft Matter. 2013 Dec 21;9(47):10.1039/C3SM51388B. doi: 10.1039/C3SM51388B. Soft Matter. 2013. PMID: 24358048 Free PMC article.
-
Overview of simulation studies on the enzymatic activity and conformational dynamics of the GTPase Ras.Mol Simul. 2014;40(10-11):839-847. doi: 10.1080/08927022.2014.895000. Epub 2014 Mar 19. Mol Simul. 2014. PMID: 26491216 Free PMC article.
-
Segregation of negatively charged phospholipids by the polycationic and farnesylated membrane anchor of Kras.Biophys J. 2010 Dec 1;99(11):3666-74. doi: 10.1016/j.bpj.2010.10.031. Biophys J. 2010. PMID: 21112291 Free PMC article.
-
RAS Nanoclusters Selectively Sort Distinct Lipid Headgroups and Acyl Chains.Front Mol Biosci. 2021 Jun 17;8:686338. doi: 10.3389/fmolb.2021.686338. eCollection 2021. Front Mol Biosci. 2021. PMID: 34222339 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

