Rice ROOT ARCHITECTURE ASSOCIATED1 binds the proteasome subunit RPT4 and is degraded in a D-box and proteasome-dependent manner
- PMID: 18701670
- PMCID: PMC2556835
- DOI: 10.1104/pp.108.125294
Rice ROOT ARCHITECTURE ASSOCIATED1 binds the proteasome subunit RPT4 and is degraded in a D-box and proteasome-dependent manner
Abstract
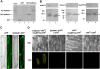
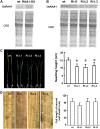
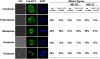
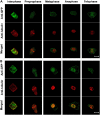
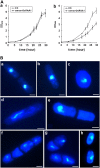
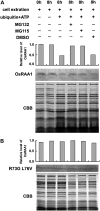
Root growth is mainly determined by cell division and subsequent elongation in the root apical area. Components regulating cell division in root meristematic cells are largely unknown. Previous studies have identified rice (Oryza sativa) ROOT ARCHITECTURE ASSOCIATED1 (OsRAA1) as a regulator in root development. Yet, the function of OsRAA1 at the cellular and molecular levels is unclear. Here, we show that OsRAA1-overexpressed transgenic rice showed reduced primary root growth, increased numbers of cells in metaphase, and reduced numbers of cells in anaphase, which suggests that OsRAA1 is responsible for limiting root growth by inhibiting the onset of anaphase. The expression of OsRAA1 in fission yeast also induced metaphase arrest, which is consistent with the fact that OsRAA1 functions through a conserved mechanism of cell cycle regulation. Moreover, a colocalization assay has shown that OsRAA1 is expressed predominantly at spindles during cell division. Yeast two-hybrid and pull-down assays, as well as a bimolecular fluorescence complementation assay, all have revealed that OsRAA1 interacts with a rice homolog of REGULATORY PARTICLE TRIPLE-A ATPASE4, a component that is involved in the ubiquitin pathway. Treating transgenic rice with specific inhibitors of the 26S proteasome blocked the degradation of OsRAA1 and increased the number of cells in metaphase. Mutation of a putative ubiquitination-targeting D-box (RGSLDLISL) in OsRAA1 interrupted the destruction of OsRAA1 in transgenic yeast. These results suggest that ubiquitination and proteasomic proteolysis are involved in OsRAA1 degradation, which is essential for the onset of anaphase, and that OsRAA1 may modulate root development mediated by the ubiquitin-proteasome pathway as a novel regulatory factor of the cell cycle.
Figures


Similar articles
-
APC-targeted RAA1 degradation mediates the cell cycle and root development in plants.Plant Signal Behav. 2010 Mar;5(3):218-23. doi: 10.4161/psb.5.3.10661. Epub 2010 Mar 14. Plant Signal Behav. 2010. PMID: 20037474 Free PMC article. Review.
-
Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity.Plant Physiol. 2004 Jul;135(3):1502-13. doi: 10.1104/pp.104.041996. Epub 2004 Jul 9. Plant Physiol. 2004. PMID: 15247372 Free PMC article.
-
The Rice E3-Ubiquitin Ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 Modulates the Expression of ROOT MEANDER CURLING, a Gene Involved in Root Mechanosensing, through the Interaction with Two ETHYLENE-RESPONSE FACTOR Transcription Factors.Plant Physiol. 2015 Nov;169(3):2275-87. doi: 10.1104/pp.15.01131. Epub 2015 Sep 17. Plant Physiol. 2015. PMID: 26381316 Free PMC article.
-
A rice really interesting new gene H2-type E3 ligase, OsSIRH2-14, enhances salinity tolerance via ubiquitin/26S proteasome-mediated degradation of salt-related proteins.Plant Cell Environ. 2019 Nov;42(11):3061-3076. doi: 10.1111/pce.13619. Epub 2019 Aug 19. Plant Cell Environ. 2019. PMID: 31325169
-
Control of metaphase-anaphase progression by proteolysis: cyclosome function regulated by the protein kinase A pathway, ubiquitination and localization.Philos Trans R Soc Lond B Biol Sci. 1999 Sep 29;354(1389):1559-69; discussion 1569-70. doi: 10.1098/rstb.1999.0499. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10582241 Free PMC article. Review.
Cited by
-
Recent progress on molecular breeding of rice in China.Plant Cell Rep. 2014 Apr;33(4):551-64. doi: 10.1007/s00299-013-1551-x. Epub 2014 Jan 19. Plant Cell Rep. 2014. PMID: 24442397 Free PMC article. Review.
-
Proofing Direct-Seeded Rice with Better Root Plasticity and Architecture.Int J Mol Sci. 2021 Jun 4;22(11):6058. doi: 10.3390/ijms22116058. Int J Mol Sci. 2021. PMID: 34199720 Free PMC article. Review.
-
UV-B irradiation-activated E3 ligase GmILPA1 modulates gibberellin catabolism to increase plant height in soybean.Nat Commun. 2023 Oct 7;14(1):6262. doi: 10.1038/s41467-023-41824-3. Nat Commun. 2023. PMID: 37805547 Free PMC article.
-
Superior Haplotypes for Early Root Vigor Traits in Rice Under Dry Direct Seeded Low Nitrogen Condition Through Genome Wide Association Mapping.Front Plant Sci. 2022 Jul 8;13:911775. doi: 10.3389/fpls.2022.911775. eCollection 2022. Front Plant Sci. 2022. PMID: 35874029 Free PMC article.
-
APC-targeted RAA1 degradation mediates the cell cycle and root development in plants.Plant Signal Behav. 2010 Mar;5(3):218-23. doi: 10.4161/psb.5.3.10661. Epub 2010 Mar 14. Plant Signal Behav. 2010. PMID: 20037474 Free PMC article. Review.
References
-
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D (1996) Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev 10 3081–3093 - PubMed
-
- Dawson SP, Arnold JE, Mayer NJ, Reynolds SE, Billett MA, Gordon C, Colleaux L, Kloetzel PM, Tanaka K, Mayer RJ (1995) Developmental changes of the 26S proteasome in abdominal intersegmental muscles of Manduca sexta during programmed cell death. J Biol Chem 270 1850–1858 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

