Arylamine N-acetyltransferases in mycobacteria
- PMID: 18680471
- PMCID: PMC2764864
- DOI: 10.2174/138920008784892100
Arylamine N-acetyltransferases in mycobacteria
Abstract
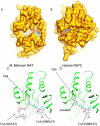
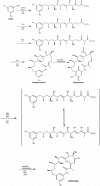
Polymorphic Human arylamine N-acetyltransferase (NAT2) inactivates the anti-tubercular drug isoniazid by acetyltransfer from acetylCoA. There are active NAT proteins encoded by homologous genes in mycobacteria including M. tuberculosis, M. bovis BCG, M. smegmatis and M. marinum. Crystallographic structures of NATs from M. smegmatis and M. marinum, as native enzymes and with isoniazid bound share a similar fold with the first NAT structure, Salmonella typhimurium NAT. There are three approximately equal domains and an active site essential catalytic triad of cysteine, histidine and aspartate in the first two domains. An acetyl group from acetylCoA is transferred to cysteine and then to the acetyl acceptor e.g. isoniazid. M. marinum NAT binds CoA in a more open mode compared with CoA binding to human NAT2. The structure of mycobacterial NAT may promote its role in synthesis of cell wall lipids, identified through gene deletion studies. NAT protein is essential for survival of M. bovis BCG in macrophage as are the proteins encoded by other genes in the same gene cluster (hsaA-D). HsaA-D degrade cholesterol, essential for mycobacterial survival inside macrophage. Nat expression remains to be fully understood but is co-ordinated with hsaA-D and other stress response genes in mycobacteria. Amide synthase genes in the streptomyces are also nat homologues. The amide synthases are predicted to catalyse intramolecular amide bond formation and creation of cyclic molecules, e.g. geldanamycin. Lack of conservation of the CoA binding cleft residues of M. marinum NAT suggests the amide synthase reaction mechanism does not involve a soluble CoA intermediate during amide formation and ring closure.
Figures


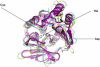


Similar articles
-
Arylamine N-acetyltransferases: from structure to function.Drug Metab Rev. 2008;40(3):479-510. doi: 10.1080/03602530802186603. Drug Metab Rev. 2008. PMID: 18642144 Review.
-
Arylamine N-acetyltransferases: a pharmacogenomic approach to drug metabolism and endogenous function.Biochem Soc Trans. 2003 Jun;31(Pt 3):615-9. doi: 10.1042/bst0310615. Biochem Soc Trans. 2003. PMID: 12773167 Review.
-
Divergence of cofactor recognition across evolution: coenzyme A binding in a prokaryotic arylamine N-acetyltransferase.J Mol Biol. 2008 Jan 4;375(1):178-91. doi: 10.1016/j.jmb.2007.10.019. Epub 2007 Oct 13. J Mol Biol. 2008. PMID: 18005984
-
Arylamine N-acetyltransferases: from drug metabolism and pharmacogenetics to drug discovery.Br J Pharmacol. 2014 Jun;171(11):2705-25. doi: 10.1111/bph.12598. Br J Pharmacol. 2014. PMID: 24467436 Free PMC article. Review.
-
Arylamine N-acetyltransferases - of mice, men and microorganisms.Trends Pharmacol Sci. 2001 Mar;22(3):140-6. doi: 10.1016/s0165-6147(00)01639-4. Trends Pharmacol Sci. 2001. PMID: 11239577 Review.
Cited by
-
cAMP-regulated protein lysine acetylases in mycobacteria.J Biol Chem. 2010 Aug 6;285(32):24313-23. doi: 10.1074/jbc.M110.118398. Epub 2010 May 27. J Biol Chem. 2010. PMID: 20507997 Free PMC article.
-
New approaches to target the mycolic acid biosynthesis pathway for the development of tuberculosis therapeutics.Curr Pharm Des. 2014;20(27):4357-78. doi: 10.2174/1381612819666131118203641. Curr Pharm Des. 2014. PMID: 24245756 Free PMC article. Review.
-
New N-acetyltransferase fold in the structure and mechanism of the phosphonate biosynthetic enzyme FrbF.J Biol Chem. 2011 Oct 14;286(41):36132-36141. doi: 10.1074/jbc.M111.263533. Epub 2011 Aug 24. J Biol Chem. 2011. PMID: 21865168 Free PMC article.
-
The Redox Cofactor F420 Protects Mycobacteria from Diverse Antimicrobial Compounds and Mediates a Reductive Detoxification System.Appl Environ Microbiol. 2016 Dec 1;82(23):6810-6818. doi: 10.1128/AEM.02500-16. Epub 2016 Sep 16. Appl Environ Microbiol. 2016. PMID: 27637879 Free PMC article.
-
Dynamic microfluidic single-cell screening identifies pheno-tuning compounds to potentiate tuberculosis therapy.Nat Commun. 2024 May 16;15(1):4175. doi: 10.1038/s41467-024-48269-2. Nat Commun. 2024. PMID: 38755132 Free PMC article.
References
-
- Tam CM, Chan SL, Kam KM, Sim E, Staples D, Sole KM, Al-Ghusein H, Mitchison DA. Int. J. Tuberc. Lung Dis. 2000;4(3):262–267. - PubMed
-
- Safdar N, Abad CL, Kaul DR, Jarrard D, Saint S. N. Eng. J. Med. 2008;358(14):1496–1501. - PubMed
-
- Sinclair JC, Sandy J, Delgoda R, Sim E, Noble MEM. Nat. Struct. Biol. 2000;7(7):560–564. - PubMed
-
- Wang H, Vath GM, Gleason KJ, Hanna PE, Wagner CR. Biochemistry. 2004;43(25):8234–8246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
