Alternative splicing and expression of human and mouse NFAT genes
- PMID: 18675896
- PMCID: PMC2577130
- DOI: 10.1016/j.ygeno.2008.06.011
Alternative splicing and expression of human and mouse NFAT genes
Abstract
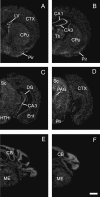
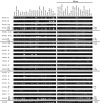
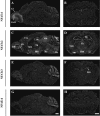
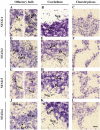
Four members of the nuclear factor of activated T cells (NFAT) family (NFATC1, NFATC2, NFATC3, and NFATC4) are Ca(2+)-regulated transcription factors that regulate several processes in vertebrates, including the development and function of the immune, cardiovascular, musculoskeletal, and nervous systems. Here we describe the structures and alternative splicing of the human and mouse NFAT genes, including novel splice variants for NFATC1, NFATC2, NFATC3, and NFATC4, and show the expression of different NFAT mRNAs in various mouse and human tissues and brain regions by RT-PCR. Our results show that alternatively spliced NFAT mRNAs are expressed differentially and could contribute to the diversity of functions of the NFAT proteins. Since NFAT family members are Ca(2+)-regulated and have critical roles in neuronal gene transcription in response to electrical activity, we describe the expression of NFATC1, NFATC2, NFATC3, and NFATC4 mRNAs in the adult mouse brain and in the adult human hippocampus using in situ hybridization and show that all NFAT mRNAs are expressed in the neurons of the mouse brain with specific patterns for each NFAT.
Figures





Similar articles
-
Regulation of different human NFAT isoforms by neuronal activity.J Neurochem. 2016 May;137(3):394-408. doi: 10.1111/jnc.13568. Epub 2016 Mar 1. J Neurochem. 2016. PMID: 26851544
-
Distinct activation properties of the nuclear factor of activated T-cells (NFAT) isoforms NFATc3 and NFATc4 in neurons.J Biol Chem. 2012 Nov 2;287(45):37594-609. doi: 10.1074/jbc.M112.365197. Epub 2012 Sep 12. J Biol Chem. 2012. PMID: 22977251 Free PMC article.
-
High extracellular calcium-induced NFATc3 regulates the expression of receptor activator of NF-κB ligand in osteoblasts.Bone. 2011 Aug;49(2):242-9. doi: 10.1016/j.bone.2011.04.006. Epub 2011 Apr 14. Bone. 2011. PMID: 21514407
-
NFAT signaling in neural development and axon growth.Int J Dev Neurosci. 2008 Apr;26(2):141-5. doi: 10.1016/j.ijdevneu.2007.10.004. Epub 2007 Nov 17. Int J Dev Neurosci. 2008. PMID: 18093786 Free PMC article. Review.
-
NFAT gene family in inflammation and cancer.Curr Mol Med. 2013 May;13(4):543-54. doi: 10.2174/1566524011313040007. Curr Mol Med. 2013. PMID: 22950383 Free PMC article. Review.
Cited by
-
NFAT/Fas signaling mediates the neuronal apoptosis and motor side effects of GSK-3 inhibition in a mouse model of lithium therapy.J Clin Invest. 2010 Jul;120(7):2432-45. doi: 10.1172/JCI37873. Epub 2010 Jun 7. J Clin Invest. 2010. PMID: 20530871 Free PMC article.
-
NFAT2 Isoforms Differentially Regulate Gene Expression, Cell Death, and Transformation through Alternative N-Terminal Domains.Mol Cell Biol. 2015 Oct 19;36(1):119-31. doi: 10.1128/MCB.00501-15. Print 2016 Jan 1. Mol Cell Biol. 2015. PMID: 26483414 Free PMC article.
-
Identification of Novel Nuclear Factor of Activated T Cell (NFAT)-associated Proteins in T Cells.J Biol Chem. 2016 Nov 11;291(46):24172-24187. doi: 10.1074/jbc.M116.739326. Epub 2016 Sep 16. J Biol Chem. 2016. PMID: 27637333 Free PMC article.
-
Genome-wide association study of CSF biomarkers Abeta1-42, t-tau, and p-tau181p in the ADNI cohort.Neurology. 2011 Jan 4;76(1):69-79. doi: 10.1212/WNL.0b013e318204a397. Epub 2010 Dec 1. Neurology. 2011. PMID: 21123754 Free PMC article.
-
Developmental expression of CREB1 and NFATC2 in pig embryos.Mol Biol Rep. 2023 Jul;50(7):6265-6271. doi: 10.1007/s11033-023-08501-6. Epub 2023 May 12. Mol Biol Rep. 2023. PMID: 37171550
References
-
- Crabtree G.R., Olson E.N. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109:S67–S79. (Suppl.) - PubMed
-
- Aramburu J., Garcia-Cozar F., Raghavan A., Okamura H., Rao A., Hogan P.G. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol. Cell. 1998;1:627–637. - PubMed
-
- Kiani A., Rao A., Aramburu J. Manipulating immune responses with immunosuppressive agents that target NFAT. Immunity. 2000;12:359–372. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

