Interaction of herpes primase with the sugar of a NTP
- PMID: 18672908
- PMCID: PMC2586337
- DOI: 10.1021/bi8008467
Interaction of herpes primase with the sugar of a NTP
Abstract
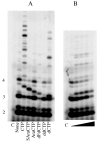
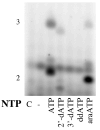
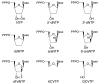
We analyzed the interaction of nucleoside triphosphates (NTPs) containing modified sugars to develop a better understanding of how DNA primase from herpes simplex virus I catalyzes primer synthesis. During the NTP binding reaction, primase tolerated a large number of modifications to the sugar ring. Altering the 2' and 3' carbons and even converting the furanose sugar into an acyclic sugar did not prevent binding. Whether or not the base on the NTP could form a correct base pair with the template base being replicated also had minimal effect on the binding reaction, indicating that primase does not use this process to discriminate between right and wrong NTPs. Rather, the key feature that primase recognizes to bind a NTP is the 5'-gamma-phosphate since converting a NTP into a NDP greatly compromised binding. During the polymerization reaction, primase tolerated substantial modification of the 2'-carbon, including the presence of either an ara or ribo hydroxyl, two hydrogens, or two fluorines. However, polymerization absolutely required that the NTP contain a 3'-hydroxyl and an intact sugar ring. Modifications at the 2'-carbon of the nucleotide at the primer 3'-terminus significantly impaired further polymerization events. Compared to a ribonucleotide, incorporation of a 2'-deoxyribo- or 2',2'-difluoro-2'-deoxyribonucleotide resulted in strong chain termination, while incorporation of an aranucleotide resulted in very strong chain termination. The implications of these data with respect to the mechanism of primase and the relationship between human and herpes primase are discussed.
Figures

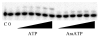
Similar articles
-
Herpes simplex virus-1 DNA primase: a remarkably inaccurate yet selective polymerase.Biochemistry. 2009 Nov 24;48(46):10866-81. doi: 10.1021/bi901476k. Biochemistry. 2009. PMID: 19835416 Free PMC article.
-
Key role of template sequence for primer synthesis by the herpes simplex virus 1 helicase-primase.Biochemistry. 2002 Dec 10;41(49):14569-79. doi: 10.1021/bi026680v. Biochemistry. 2002. PMID: 12463757
-
Mechanism of primer synthesis by the herpes simplex virus 1 helicase-primase.Biochemistry. 2004 Feb 17;43(6):1754-62. doi: 10.1021/bi035519x. Biochemistry. 2004. PMID: 14769053
-
The two helicases of herpes simplex virus type 1 (HSV-1).Front Biosci. 2006 Sep 1;11:2213-23. doi: 10.2741/1964. Front Biosci. 2006. PMID: 16720308 Review.
-
Mechanism of DNA primer synthesis by human PrimPol.Enzymes. 2019;45:289-310. doi: 10.1016/bs.enz.2019.06.003. Epub 2019 Jul 13. Enzymes. 2019. PMID: 31627881 Review.
Cited by
-
Herpes simplex virus-1 helicase-primase: roles of each subunit in DNA binding and phosphodiester bond formation.Biochemistry. 2009 Nov 3;48(43):10199-207. doi: 10.1021/bi9010144. Biochemistry. 2009. PMID: 19788334 Free PMC article.
-
Initiation of new DNA strands by the herpes simplex virus-1 primase-helicase complex and either herpes DNA polymerase or human DNA polymerase alpha.J Biol Chem. 2009 Jan 16;284(3):1523-32. doi: 10.1074/jbc.M805476200. Epub 2008 Nov 20. J Biol Chem. 2009. PMID: 19028696 Free PMC article.
-
Mechanisms by which human DNA primase chooses to polymerize a nucleoside triphosphate.Biochemistry. 2010 Feb 2;49(4):727-35. doi: 10.1021/bi9019516. Biochemistry. 2010. PMID: 20030400 Free PMC article.
-
Coordinated leading and lagging strand DNA synthesis by using the herpes simplex virus 1 replication complex and minicircle DNA templates.J Virol. 2011 Jan;85(2):957-67. doi: 10.1128/JVI.01688-10. Epub 2010 Nov 10. J Virol. 2011. PMID: 21068232 Free PMC article.
-
Structures to complement the archaeo-eukaryotic primases catalytic cycle description: What's next?Comput Struct Biotechnol J. 2015 May 2;13:339-51. doi: 10.1016/j.csbj.2015.04.006. eCollection 2015. Comput Struct Biotechnol J. 2015. PMID: 25987967 Free PMC article. Review.
References
-
- Boehmer PE, Lehman IR. Herpes Simplex Virus DNA Replication. Annu Rev Biochem. 1997;66:347–384. - PubMed
-
- Crute JJ, Ctygon CA, Hargrave KD, Simoneau B, Faucher AM, Bolger G, Kibler P, Liuzzi M, Cordingley MG. Herpes Simplex Virus Helicase-primase Inhibitors Are Active in Animal Models of Human Disease. Nature Medicine. 2002;8:386–391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

