Autoregulation of Th1-mediated inflammation by twist1
- PMID: 18663125
- PMCID: PMC2525589
- DOI: 10.1084/jem.20072468
Autoregulation of Th1-mediated inflammation by twist1
Abstract
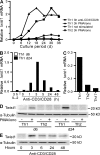
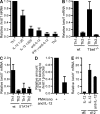
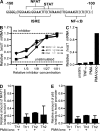
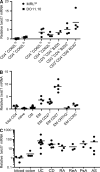
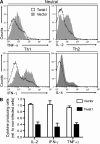
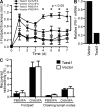
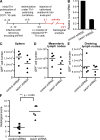
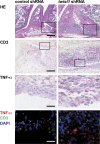
The basic helix-loop-helix transcriptional repressor twist1, as an antagonist of nuclear factor kappaB (NF-kappaB)-dependent cytokine expression, is involved in the regulation of inflammation-induced immunopathology. We show that twist1 is expressed by activated T helper (Th) 1 effector memory (EM) cells. Induction of twist1 in Th cells depended on NF-kappaB, nuclear factor of activated T cells (NFAT), and interleukin (IL)-12 signaling via signal transducer and activator of transcription (STAT) 4. Expression of twist1 was transient after T cell receptor engagement, and increased upon repeated stimulation of Th1 cells. Imprinting for enhanced twist1 expression was characteristic of repeatedly restimulated EM Th cells, and thus of the pathogenic memory Th cells characteristic of chronic inflammation. Th lymphocytes from the inflamed joint or gut tissue of patients with rheumatic diseases, Crohn's disease or ulcerative colitis expressed high levels of twist1. Expression of twist1 in Th1 lymphocytes limited the expression of the cytokines interferon-gamma, IL-2, and tumor necrosis factor-alpha, and ameliorated Th1-mediated immunopathology in delayed-type hypersensitivity and antigen-induced arthritis.
Figures


Similar articles
-
[The pro-inflammatory immunological memory : twist1 as a marker for chronically activated T lymphocytes].Z Rheumatol. 2008 Dec;67(8):684-8. doi: 10.1007/s00393-008-0403-5. Z Rheumatol. 2008. PMID: 19002471 German.
-
miR-148a is upregulated by Twist1 and T-bet and promotes Th1-cell survival by regulating the proapoptotic gene Bim.Eur J Immunol. 2015 Apr;45(4):1192-205. doi: 10.1002/eji.201444633. Epub 2015 Jan 22. Eur J Immunol. 2015. PMID: 25486906 Free PMC article.
-
Regulation of Fatty Acid Oxidation by Twist 1 in the Metabolic Adaptation of T Helper Lymphocytes to Chronic Inflammation.Arthritis Rheumatol. 2019 Oct;71(10):1756-1765. doi: 10.1002/art.40939. Epub 2019 Sep 5. Arthritis Rheumatol. 2019. PMID: 31131995 Free PMC article.
-
The emerging role of Twist proteins in hematopoietic cells and hematological malignancies.Blood Cancer J. 2014 Apr 25;4(4):e206. doi: 10.1038/bcj.2014.22. Blood Cancer J. 2014. PMID: 24769647 Free PMC article. Review.
-
[How T lymphocytes coordinate rheumatic inflammation].Z Rheumatol. 2022 Oct;81(8):635-641. doi: 10.1007/s00393-022-01186-5. Epub 2022 Apr 5. Z Rheumatol. 2022. PMID: 35380250 Review. German.
Cited by
-
A Multi-Omics Analysis of Mucosal-Associated-Invariant T Cells Reveals Key Drivers of Distinct Modes of Activation.Front Immunol. 2021 May 24;12:616967. doi: 10.3389/fimmu.2021.616967. eCollection 2021. Front Immunol. 2021. PMID: 34108957 Free PMC article.
-
Novel therapies for memory cells in autoimmune diseases.Clin Exp Immunol. 2015 Jun;180(3):353-60. doi: 10.1111/cei.12602. Epub 2015 Apr 23. Clin Exp Immunol. 2015. PMID: 25682849 Free PMC article. Review.
-
Circadian Pharmacological Effects of Paeoniflorin on Mice With Urticaria-like Lesions.Front Pharmacol. 2022 Feb 9;12:639580. doi: 10.3389/fphar.2021.639580. eCollection 2021. Front Pharmacol. 2022. PMID: 35222003 Free PMC article.
-
EZH2 inhibition suppresses endometrial cancer progression via miR-361/Twist axis.Oncotarget. 2017 Feb 21;8(8):13509-13520. doi: 10.18632/oncotarget.14586. Oncotarget. 2017. PMID: 28088786 Free PMC article.
-
Twist1 regulates Ifng expression in Th1 cells by interfering with Runx3 function.J Immunol. 2012 Jul 15;189(2):832-40. doi: 10.4049/jimmunol.1200854. Epub 2012 Jun 8. J Immunol. 2012. PMID: 22685315 Free PMC article.
References
-
- Chen, Z.F., and R.R. Behringer. 1995. Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 9:686–699. - PubMed
-
- Yang, J., S.A. Mani, J.L. Donaher, S. Ramaswamy, R.A. Itzykson, C. Come, P. Savagner, I. Gitelman, A. Richardson, and R.A. Weinberg. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117:927–939. - PubMed
-
- Sosic, D., J.A. Richardson, K. Yu, D.M. Ornitz, and E.N. Olson. 2003. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 112:169–180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

