Mechanistic insights into phosphoprotein-binding FHA domains
- PMID: 18656966
- PMCID: PMC2962622
- DOI: 10.1021/ar700148u
Mechanistic insights into phosphoprotein-binding FHA domains
Abstract
[Structure: see text]. FHA domains are protein modules that switch signals in diverse biological pathways by monitoring the phosphorylation of threonine residues of target proteins. As part of the effort to gain insight into cellular avoidance of cancer, FHA domains involved in the cellular response to DNA damage have been especially well-characterized. The complete protein where the FHA domain resides and the interaction partners determine the nature of the signaling. Thus, a key biochemical question is how do FHA domains pick out their partners from among thousands of alternatives in the cell? This Account discusses the structure, affinity, and specificity of FHA domains and the formation of their functional structure. Although FHA domains share sequence identity at only five loop residues, they all fold into a beta-sandwich of two beta-sheets. The conserved arginine and serine of the recognition loops recognize the phosphorylation of the threonine targeted. Side chains emanating from loops that join beta-strand 4 with 5, 6 with 7, or 10 with 11 make specific contacts with amino acids of the ligand that tailor sequence preferences. Many FHA domains choose a partner in extended conformation, somewhat according to the residue three after the phosphothreonine in sequence (pT + 3 position). One group of FHA domains chooses a short carboxylate-containing side chain at pT + 3. Another group chooses a long, branched aliphatic side chain. A third group prefers other hydrophobic or uncharged polar side chains at pT + 3. However, another FHA domain instead chooses on the basis of pT - 2, pT - 3, and pT + 1 positions. An FHA domain from a marker of human cancer instead chooses a much longer protein fragment that adds a beta-strand to its beta-sheet and that presents hydrophobic residues from a novel helix to the usual recognition surface. This novel recognition site and more remote sites for the binding of other types of protein partners were predicted for the entire family of FHA domains by a bioinformatics approach. The phosphopeptide-dependent dynamics of an FHA domain, SH2 domain, and PTB domain suggest a common theme: rigid, preformed binding surfaces support van der Waals contacts that provide favorable binding enthalpy. Despite the lack of pronounced conformational changes in FHA domains linked to binding events, more subtle adjustments may be possible. In the one FHA domain tested, phosphothreonine peptide binding is accompanied by increased flexibility just outside the binding site and increased rigidity across the beta-sandwich. The folding of the same FHA domain progresses through near-native intermediates that stabilize the recognition loops in the center of the phosphoprotein-binding surface; this may promote rigidity in the interface and affinity for targets phosphorylated on threonine.
Figures

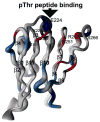
Similar articles
-
Achieving peptide binding specificity and promiscuity by loops: case of the forkhead-associated domain.PLoS One. 2014 May 28;9(5):e98291. doi: 10.1371/journal.pone.0098291. eCollection 2014. PLoS One. 2014. PMID: 24870410 Free PMC article.
-
PhosphoThr peptide binding globally rigidifies much of the FHA domain from Arabidopsis receptor kinase-associated protein phosphatase.Biochemistry. 2005 Aug 2;44(30):10119-34. doi: 10.1021/bi050414a. Biochemistry. 2005. PMID: 16042389 Free PMC article.
-
NMR structure of the forkhead-associated domain from the Arabidopsis receptor kinase-associated protein phosphatase.Proc Natl Acad Sci U S A. 2003 Sep 30;100(20):11261-6. doi: 10.1073/pnas.2031918100. Epub 2003 Sep 18. Proc Natl Acad Sci U S A. 2003. PMID: 14500786 Free PMC article.
-
Structure and function of the phosphothreonine-specific FHA domain.Sci Signal. 2008 Dec 23;1(51):re12. doi: 10.1126/scisignal.151re12. Sci Signal. 2008. PMID: 19109241 Review.
-
The FHA domain mediates phosphoprotein interactions.J Cell Sci. 2000 Dec;113 Pt 23:4143-9. doi: 10.1242/jcs.113.23.4143. J Cell Sci. 2000. PMID: 11069759 Review.
Cited by
-
Physicochemical mechanisms of protein regulation by phosphorylation.Front Genet. 2014 Aug 7;5:270. doi: 10.3389/fgene.2014.00270. eCollection 2014. Front Genet. 2014. PMID: 25147561 Free PMC article. Review.
-
Achieving peptide binding specificity and promiscuity by loops: case of the forkhead-associated domain.PLoS One. 2014 May 28;9(5):e98291. doi: 10.1371/journal.pone.0098291. eCollection 2014. PLoS One. 2014. PMID: 24870410 Free PMC article.
-
Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases.Proc Natl Acad Sci U S A. 2010 Apr 20;107(16):7521-6. doi: 10.1073/pnas.0913482107. Epub 2010 Apr 5. Proc Natl Acad Sci U S A. 2010. PMID: 20368441 Free PMC article.
-
Specific recognition of a multiply phosphorylated motif in the DNA repair scaffold XRCC1 by the FHA domain of human PNK.Nucleic Acids Res. 2009 Apr;37(5):1701-12. doi: 10.1093/nar/gkn1086. Epub 2009 Jan 20. Nucleic Acids Res. 2009. PMID: 19155274 Free PMC article.
-
The intrinsically disordered N-terminal region of AtREM1.3 remorin protein mediates protein-protein interactions.J Biol Chem. 2012 Nov 16;287(47):39982-91. doi: 10.1074/jbc.M112.414292. Epub 2012 Oct 1. J Biol Chem. 2012. PMID: 23027878 Free PMC article.
References
-
- Westheimer FH. Why nature chose phosphates. Science. 1987;235(4793):1173–8. - PubMed
-
- Yaffe MB, Cantley LC. Grabbing phosphoproteins. Science. 1999;402:30–31. - PubMed
-
- Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7(7):473–83. - PubMed
-
- Yaffe MB, Smerdon SJ. The use of in vitro peptide-library screens in the analysis of phosphoserine/threonine-binding domain structure and function. Annu Rev Biophys Biomol Struct. 2004;33:225–44. - PubMed
-
- Durocher D, Jackson SP. The FHA domain. FEBS Lett. 2002;513(1):58–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

