A new role for HIV nucleocapsid protein in modulating the specificity of plus strand priming
- PMID: 18632127
- PMCID: PMC2607142
- DOI: 10.1016/j.virol.2008.06.002
A new role for HIV nucleocapsid protein in modulating the specificity of plus strand priming
Abstract
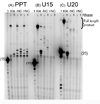
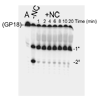
The current study indicates a new role for HIV nucleocapsid protein (NC) in modulating the specificity of plus strand priming. RNase H cleavage by reverse transcriptase (RT) during minus strand synthesis gives rise to RNA fragments that could potentially be used as primers for synthesis of the plus strand, leading to the initiation of priming from multiple points as has been observed for other retroviruses. For HIV, the central and 3' polypurine tracts (PPTs) are the major sites of plus strand initiation. Using reconstituted in vitro assays, results showed that NC greatly reduced the efficiency of extension of non-PPT RNA primers, but not PPT. Experiments mimicking HIV replication showed that RT generated and used both PPT and non-PPT RNAs to initiate "plus strand" synthesis, but non-PPT usage was strongly inhibited by NC. The results support a role for NC in specifying primer usage during plus strand synthesis.
Figures





Similar articles
-
Fidelity of plus-strand priming requires the nucleic acid chaperone activity of HIV-1 nucleocapsid protein.Nucleic Acids Res. 2009 Apr;37(6):1755-66. doi: 10.1093/nar/gkn1045. Epub 2009 Jan 21. Nucleic Acids Res. 2009. PMID: 19158189 Free PMC article.
-
RNA degradation and primer selection by Moloney murine leukemia virus reverse transcriptase contribute to the accuracy of plus strand initiation.J Biol Chem. 2000 Apr 28;275(17):13061-70. doi: 10.1074/jbc.275.17.13061. J Biol Chem. 2000. PMID: 10777611
-
Use of an oligoribonucleotide containing the polypurine tract sequence as a primer by HIV reverse transcriptase.J Biol Chem. 1995 Nov 24;270(47):28169-76. doi: 10.1074/jbc.270.47.28169. J Biol Chem. 1995. PMID: 7499308
-
'Binding, bending and bonding': polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus.Int J Biochem Cell Biol. 2004 Sep;36(9):1752-66. doi: 10.1016/j.biocel.2004.02.016. Int J Biochem Cell Biol. 2004. PMID: 15183342 Review.
-
Interaction of retroviral reverse transcriptase with template-primer duplexes during replication.Prog Nucleic Acid Res Mol Biol. 1998;58:339-93. doi: 10.1016/s0079-6603(08)60041-0. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9308371 Review.
Cited by
-
Comparative nucleic acid chaperone properties of the nucleocapsid protein NCp7 and Tat protein of HIV-1.Virus Res. 2012 Nov;169(2):349-60. doi: 10.1016/j.virusres.2012.06.021. Epub 2012 Jun 26. Virus Res. 2012. PMID: 22743066 Free PMC article. Review.
-
Specific implications of the HIV-1 nucleocapsid zinc fingers in the annealing of the primer binding site complementary sequences during the obligatory plus strand transfer.Nucleic Acids Res. 2011 Aug;39(15):6633-45. doi: 10.1093/nar/gkr274. Epub 2011 May 4. Nucleic Acids Res. 2011. PMID: 21543454 Free PMC article.
-
Nucleocapsid protein annealing of a primer-template enhances (+)-strand DNA synthesis and fidelity by HIV-1 reverse transcriptase.J Mol Biol. 2012 Feb 3;415(5):866-80. doi: 10.1016/j.jmb.2011.12.034. Epub 2011 Dec 23. J Mol Biol. 2012. PMID: 22210155 Free PMC article.
-
A protein ballet around the viral genome orchestrated by HIV-1 reverse transcriptase leads to an architectural switch: from nucleocapsid-condensed RNA to Vpr-bridged DNA.Virus Res. 2013 Feb;171(2):287-303. doi: 10.1016/j.virusres.2012.09.008. Epub 2012 Sep 24. Virus Res. 2013. PMID: 23017337 Free PMC article.
-
An Evolutionary/Biochemical Connection between Promoter- and Primer-Dependent Polymerases Revealed by Systematic Evolution of Ligands by Exponential Enrichment.J Bacteriol. 2018 Mar 12;200(7):e00579-17. doi: 10.1128/JB.00579-17. Print 2018 Apr 1. J Bacteriol. 2018. PMID: 29339418 Free PMC article.
References
-
- Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000a;301(2):491–511. - PubMed
-
- Amarasinghe GK, De Guzman RN, Turner RB, Summers MF. NMR structure of stem-loop SL2 of the HIV-1 psi RNA packaging signal reveals a novel A-U-A base-triple platform. J. Mol. Biol. 2000b;299(1):145–156. - PubMed
-
- Baba S, Takahashi K, Koyanagi Y, Yamamoto N, Takaku H, Gorelick RJ, Kawai G. Role of the zinc fingers of HIV-1 nucleocapsid protein in maturation of genomic RNA. J. Biochem. (Tokyo) 2003;134(5):637–639. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

