The protein kinase C-responsive inhibitory domain of CARD11 functions in NF-kappaB activation to regulate the association of multiple signaling cofactors that differentially depend on Bcl10 and MALT1 for association
- PMID: 18625728
- PMCID: PMC2546934
- DOI: 10.1128/MCB.00418-08
The protein kinase C-responsive inhibitory domain of CARD11 functions in NF-kappaB activation to regulate the association of multiple signaling cofactors that differentially depend on Bcl10 and MALT1 for association
Abstract
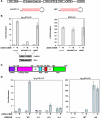
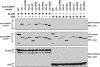
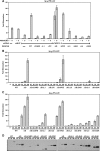
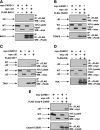
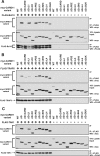
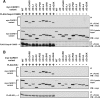
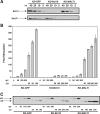
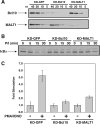
The activation of NF-kappaB by T-cell receptor (TCR) signaling is critical for T-cell activation during the adaptive immune response. CARD11 is a multidomain adapter that is required for TCR signaling to the IkappaB kinase (IKK) complex. During TCR signaling, the region in CARD11 between the coiled-coil and PDZ domains is phosphorylated by protein kinase Ctheta (PKCtheta) in a required step in NF-kappaB activation. In this report, we demonstrate that this region functions as an inhibitory domain (ID) that controls the association of CARD11 with multiple signaling cofactors, including Bcl10, TRAF6, TAK1, IKKgamma, and caspase-8, through an interaction that requires both the caspase recruitment domain (CARD) and the coiled-coil domain. Consistent with the ID-mediated control of their association, we demonstrate that TRAF6 and caspase-8 associate with CARD11 in T cells in a signal-inducible manner. Using an RNA interference rescue assay, we demonstrate that the CARD, linker 1, coiled-coil, linker 3, SH3, linker 4, and GUK domains are each required for TCR signaling to NF-kappaB downstream of ID neutralization. Requirements for the CARD, linker 1, and coiled-coil domains in signaling are consistent with their roles in the association of CARD11 with Bcl10, TRAF6, TAK1, caspase-8, and IKKgamma. Using Bcl10- and MALT1-deficient cells, we show that CARD11 can recruit signaling cofactors independently of one another in a signal-inducible manner.
Figures






Similar articles
-
Molecular Determinants of Scaffold-induced Linear Ubiquitinylation of B Cell Lymphoma/Leukemia 10 (Bcl10) during T Cell Receptor and Oncogenic Caspase Recruitment Domain-containing Protein 11 (CARD11) Signaling.J Biol Chem. 2016 Dec 9;291(50):25921-25936. doi: 10.1074/jbc.M116.754028. Epub 2016 Oct 24. J Biol Chem. 2016. PMID: 27777308 Free PMC article.
-
Protein kinase C-δ negatively regulates T cell receptor-induced NF-κB activation by inhibiting the assembly of CARMA1 signalosome.J Biol Chem. 2012 Jun 8;287(24):20081-7. doi: 10.1074/jbc.M111.335463. Epub 2012 Apr 23. J Biol Chem. 2012. PMID: 22528498 Free PMC article.
-
A quantitative signaling screen identifies CARD11 mutations in the CARD and LATCH domains that induce Bcl10 ubiquitination and human lymphoma cell survival.Mol Cell Biol. 2013 Jan;33(2):429-43. doi: 10.1128/MCB.00850-12. Epub 2012 Nov 12. Mol Cell Biol. 2013. PMID: 23149938 Free PMC article.
-
Genetic errors of the human caspase recruitment domain-B-cell lymphoma 10-mucosa-associated lymphoid tissue lymphoma-translocation gene 1 (CBM) complex: Molecular, immunologic, and clinical heterogeneity.J Allergy Clin Immunol. 2015 Nov;136(5):1139-49. doi: 10.1016/j.jaci.2015.06.031. Epub 2015 Aug 12. J Allergy Clin Immunol. 2015. PMID: 26277595 Free PMC article. Review.
-
The CARD11-BCL10-MALT1 (CBM) signalosome complex: Stepping into the limelight of human primary immunodeficiency.J Allergy Clin Immunol. 2014 Aug;134(2):276-84. doi: 10.1016/j.jaci.2014.06.015. J Allergy Clin Immunol. 2014. PMID: 25087226 Free PMC article. Review.
Cited by
-
BCL10-CARD11 Fusion Mimics an Active CARD11 Seed That Triggers Constitutive BCL10 Oligomerization and Lymphocyte Activation.Front Immunol. 2018 Nov 20;9:2695. doi: 10.3389/fimmu.2018.02695. eCollection 2018. Front Immunol. 2018. PMID: 30515170 Free PMC article.
-
CARMA3: Scaffold Protein Involved in NF-κB Signaling.Front Immunol. 2019 Feb 13;10:176. doi: 10.3389/fimmu.2019.00176. eCollection 2019. Front Immunol. 2019. PMID: 30814996 Free PMC article. Review.
-
Gain-of-function mutations and immunodeficiency: at a loss for proper tuning of lymphocyte signaling.Curr Opin Allergy Clin Immunol. 2015 Dec;15(6):533-8. doi: 10.1097/ACI.0000000000000217. Curr Opin Allergy Clin Immunol. 2015. PMID: 26406182 Free PMC article. Review.
-
L-CBM signaling in lymphocyte development and function.J Blood Med. 2010;1:93-104. doi: 10.2147/JBM.S9772. Epub 2010 Jun 4. J Blood Med. 2010. PMID: 22282688 Free PMC article.
-
Identification and Characterization of a Germline Mutation in CARD11 From a Chinese Case of B Cell Expansion With NF-κB and T Cell Anergy.Front Immunol. 2021 Sep 7;12:676386. doi: 10.3389/fimmu.2021.676386. eCollection 2021. Front Immunol. 2021. PMID: 34557185 Free PMC article.
References
-
- Akagi, T., M. Motegi, A. Tamura, R. Suzuki, Y. Hosokawa, H. Suzuki, H. Ota, S. Nakamura, Y. Morishima, M. Taniwaki, and M. Seto. 1999. A novel gene, MALT1 at 18q21, is involved in t(11;18) (q21;q21) found in low-grade B-cell lymphoma of mucosa-associated lymphoid tissue. Oncogene 185785-5794. - PubMed
-
- Bell, S., K. Degitz, M. Quirling, N. Jilg, S. Page, and K. Brand. 2003. Involvement of NF-κB signalling in skin physiology and disease. Cell Signal. 151-7. - PubMed
-
- Bertin, J., L. Wang, Y. Guo, M. D. Jacobson, J. L. Poyet, S. M. Srinivasula, S. Merriam, P. S. DiStefano, and E. S. Alnemri. 2001. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-κB. J. Biol. Chem. 27611877-11882. - PubMed
-
- Bidere, N., A. L. Snow, K. Sakai, L. Zheng, and M. J. Lenardo. 2006. Caspase-8 regulation by direct interaction with TRAF6 in T cell receptor-induced NF-κB activation. Curr. Biol. 161666-1671. - PubMed
-
- Cannons, J. L., L. J. Yu, B. Hill, L. A. Mijares, D. Dombroski, K. E. Nichols, A. Antonellis, G. A. Koretzky, K. Gardner, and P. L. Schwartzberg. 2004. SAP regulates TH2 differentiation and PKC-θ-mediated activation of NF-κB1. Immunity 21693-706. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
