A combinatorial H4 tail library for exploring the histone code
- PMID: 18616348
- PMCID: PMC2614903
- DOI: 10.1021/bi800766k
A combinatorial H4 tail library for exploring the histone code
Abstract
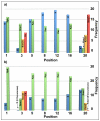
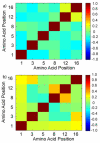
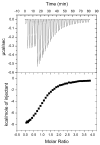
Histone modifications modulate chromatin structure and function. A posttranslational modification-randomized, combinatorial library based on the first 21 residues of histone H4 was designed for systematic examination of proteins that interpret a histone code. The 800-member library represented all permutations of most known modifications within the N-terminal tail of histone H4. To determine its utility in a protein binding assay, the on-bead library was screened with an antibody directed against phosphoserine 1 of H4. Among the hits, 59 of 60 sequences were phosphorylated at S1, while 30 of 30 of those selected from the nonhits were unphosphorylated. A 512-member version of the library was then used to determine the binding specificity of the double tudor domain of hJMJD2A, a histone demethylase involved in transcriptional repression. Global linear least-squares fitting of modifications from the identified peptides (40 hits and 34 nonhits) indicated that methylation of K20 was the primary determinant for binding, but that phosphorylation and acetylation of neighboring sites attenuated the interaction. To validate the on-bead screen, isothermal titration calorimetry was performed with 13 H4 peptides. Dissociation constants ranged from 1 mM to 1 microM and corroborated the screening results. The general approach should be useful for probing the specificity of any histone-binding protein.
Figures
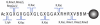
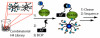

Similar articles
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Tobacco Product Use and Associated Factors Among Middle and High School Students - National Youth Tobacco Survey, United States, 2021.MMWR Surveill Summ. 2022 Mar 11;71(5):1-29. doi: 10.15585/mmwr.ss7105a1. MMWR Surveill Summ. 2022. PMID: 35271557 Free PMC article.
-
Interventions for supporting pregnant women's decision-making about mode of birth after a caesarean.Cochrane Database Syst Rev. 2013 Jul 30;2013(7):CD010041. doi: 10.1002/14651858.CD010041.pub2. Cochrane Database Syst Rev. 2013. PMID: 23897547 Free PMC article. Review.
Cited by
-
Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9.J Biol Chem. 2011 Apr 1;286(13):11779-91. doi: 10.1074/jbc.M110.208207. Epub 2011 Jan 28. J Biol Chem. 2011. PMID: 21278251 Free PMC article.
-
Select human cancer mutants of NRMT1 alter its catalytic activity and decrease N-terminal trimethylation.Protein Sci. 2017 Aug;26(8):1639-1652. doi: 10.1002/pro.3202. Epub 2017 Jun 11. Protein Sci. 2017. PMID: 28556566 Free PMC article.
-
Chemical Biology Approaches to Identify and Profile Interactors of Chromatin Modifications.ACS Chem Biol. 2023 Apr 21;18(4):1014-1026. doi: 10.1021/acschembio.1c00794. Epub 2022 Mar 3. ACS Chem Biol. 2023. PMID: 35238546 Free PMC article. Review.
-
Chemical approaches to understand the language of histone modifications.ACS Chem Biol. 2011 Oct 21;6(10):987-99. doi: 10.1021/cb200142c. Epub 2011 Aug 17. ACS Chem Biol. 2011. PMID: 21827195 Free PMC article. Review.
-
Histone modifications and a choice of variant: a language that helps the genome express itself.F1000Prime Rep. 2014 Sep 4;6:76. doi: 10.12703/P6-76. eCollection 2014. F1000Prime Rep. 2014. PMID: 25343033 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

