The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer
- PMID: 18596042
- PMCID: PMC2528990
- DOI: 10.1074/jbc.M803370200
The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer
Abstract
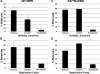
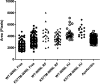
Werner syndrome is an inherited disease displaying a premature aging phenotype. The gene mutated in Werner syndrome encodes both a 3' --> 5' DNA helicase and a 3' --> 5' DNA exonuclease. Both WRN helicase and exonuclease preferentially utilize DNA substrates containing alternate secondary structures. By virtue of its ability to resolve such DNA structures, WRN is postulated to prevent the stalling and collapse of replication forks that encounter damaged DNA. Using electron microscopy, we visualized the binding of full-length WRN to DNA templates containing replication forks and Holliday junctions, intermediates observed during DNA replication and recombination, respectively. We show that both wild-type WRN and a helicase-defective mutant bind with exceptionally high specificity (>1000-fold) to DNA secondary structures at the replication fork and at Holliday junctions. Little or no binding is observed elsewhere on the DNA molecules. Calculations of the molecular weight of full-length WRN revealed that, in solution, WRN exists predominantly as a dimer. However, WRN bound to DNA is larger; the mass is consistent with that of a tetramer.
Figures



Similar articles
-
Replication fork regression in vitro by the Werner syndrome protein (WRN): holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity.Nucleic Acids Res. 2007;35(17):5729-47. doi: 10.1093/nar/gkm561. Epub 2007 Aug 23. Nucleic Acids Res. 2007. PMID: 17717003 Free PMC article.
-
The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) holliday junctions to functional replication forks.Biochemistry. 2011 Aug 16;50(32):6774-88. doi: 10.1021/bi2001054. Epub 2011 Jul 21. Biochemistry. 2011. PMID: 21736299 Free PMC article.
-
WRN helicase defective in the premature aging disorder Werner syndrome genetically interacts with topoisomerase 3 and restores the top3 slow growth phenotype of sgs1 top3.Aging (Albany NY). 2009 Feb 5;1(2):219-33. doi: 10.18632/aging.100020. Aging (Albany NY). 2009. PMID: 20157511 Free PMC article.
-
The Werner syndrome protein: linking the replication checkpoint response to genome stability.Aging (Albany NY). 2011 Mar;3(3):311-8. doi: 10.18632/aging.100293. Aging (Albany NY). 2011. PMID: 21389352 Free PMC article. Review.
-
Roles of the Werner syndrome RecQ helicase in DNA replication.DNA Repair (Amst). 2008 Nov 1;7(11):1776-86. doi: 10.1016/j.dnarep.2008.07.017. Epub 2008 Sep 6. DNA Repair (Amst). 2008. PMID: 18722555 Free PMC article. Review.
Cited by
-
Distinct roles of RECQ1 in the maintenance of genomic stability.DNA Repair (Amst). 2010 Mar 2;9(3):315-24. doi: 10.1016/j.dnarep.2009.12.010. Epub 2010 Jan 12. DNA Repair (Amst). 2010. PMID: 20061189 Free PMC article. Review.
-
WRN participates in translesion synthesis pathway through interaction with NBS1.Mech Ageing Dev. 2010 Jun;131(6):436-44. doi: 10.1016/j.mad.2010.06.005. Epub 2010 Jun 17. Mech Ageing Dev. 2010. PMID: 20600238 Free PMC article.
-
Dynamics of the DNA repair proteins WRN and BLM in the nucleoplasm and nucleoli.Eur Biophys J. 2014 Nov;43(10-11):509-16. doi: 10.1007/s00249-014-0981-x. Epub 2014 Aug 14. Eur Biophys J. 2014. PMID: 25119658 Free PMC article.
-
DNA repair and replication fork helicases are differentially affected by alkyl phosphotriester lesion.J Biol Chem. 2012 Jun 1;287(23):19188-98. doi: 10.1074/jbc.M112.352757. Epub 2012 Apr 12. J Biol Chem. 2012. PMID: 22500020 Free PMC article.
-
Werner syndrome protein works as a dimer for unwinding and replication fork regression.Nucleic Acids Res. 2023 Jan 11;51(1):337-348. doi: 10.1093/nar/gkac1200. Nucleic Acids Res. 2023. PMID: 36583333 Free PMC article.
References
-
- Singleton, M. R., Dillingham, M. S., and Wigley, D. B. (2007) Annu. Rev. Biochem. 7623 –50 - PubMed
-
- Ozgenc, A., and Loeb, L. A. (2005) Mutat. Res. 577237 –251 - PubMed
-
- Kamath-Loeb, A. S., Fry, M., and Loeb, L. A. (2006) DNA Helicases and Human Disease, pp.435 –460, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

