Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5
- PMID: 18591409
- PMCID: PMC2442638
- DOI: 10.1084/jem.20080091
Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5
Abstract
The ribonucleic acid (RNA) helicases retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) recognize distinct viral and synthetic RNAs, leading to the production of interferons. Although 5'-triphosphate single-stranded RNA is a RIG-I ligand, the role of RIG-I and MDA5 in double-stranded (ds) RNA recognition remains to be characterized. In this study, we show that the length of dsRNA is important for differential recognition by RIG-I and MDA5. The MDA5 ligand, polyinosinic-polycytidylic acid, was converted to a RIG-I ligand after shortening of the dsRNA length. In addition, viral dsRNAs differentially activated RIG-I and MDA5, depending on their length. Vesicular stomatitis virus infection generated dsRNA, which is responsible for RIG-I-mediated recognition. Collectively, RIG-I detects dsRNAs without a 5'-triphosphate end, and RIG-I and MDA5 selectively recognize short and long dsRNAs, respectively.
Figures


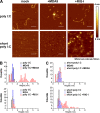
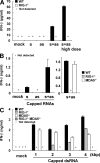
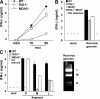
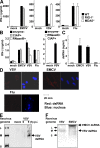
Comment in
-
Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity.J Exp Med. 2008 Jul 7;205(7):1523-7. doi: 10.1084/jem.20081210. J Exp Med. 2008. PMID: 18591413 Free PMC article.
Similar articles
-
Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity.J Exp Med. 2008 Jul 7;205(7):1523-7. doi: 10.1084/jem.20081210. J Exp Med. 2008. PMID: 18591413 Free PMC article.
-
Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses.Nature. 2006 May 4;441(7089):101-5. doi: 10.1038/nature04734. Epub 2006 Apr 9. Nature. 2006. PMID: 16625202
-
SerpinB9 expression in human renal tubular epithelial cells is induced by triggering of the viral dsRNA sensors TLR3, MDA5 and RIG-I.Nephrol Dial Transplant. 2012 Jul;27(7):2746-54. doi: 10.1093/ndt/gfr690. Epub 2011 Dec 13. Nephrol Dial Transplant. 2012. PMID: 22167597
-
Proofreading mechanisms of the innate immune receptor RIG-I: distinguishing self and viral RNA.Biochem Soc Trans. 2024 Jun 26;52(3):1131-1148. doi: 10.1042/BST20230724. Biochem Soc Trans. 2024. PMID: 38884803 Free PMC article. Review.
-
MDA5-filament, dynamics and disease.Curr Opin Virol. 2015 Jun;12:20-5. doi: 10.1016/j.coviro.2015.01.011. Epub 2015 Feb 9. Curr Opin Virol. 2015. PMID: 25676875 Free PMC article. Review.
Cited by
-
MDA5 Is a Major Determinant of Developing Symptoms in Critically Ill COVID-19 Patients.Clin Rev Allergy Immunol. 2024 Dec;67(1-3):58-72. doi: 10.1007/s12016-024-09008-z. Epub 2024 Oct 26. Clin Rev Allergy Immunol. 2024. PMID: 39460899 Review.
-
Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity.PLoS Pathog. 2013;9(4):e1003298. doi: 10.1371/journal.ppat.1003298. Epub 2013 Apr 25. PLoS Pathog. 2013. PMID: 23633948 Free PMC article.
-
DUSP4 modulates RIG-I- and STING-mediated IRF3-type I IFN response.Cell Death Differ. 2024 Mar;31(3):280-291. doi: 10.1038/s41418-024-01269-7. Epub 2024 Feb 21. Cell Death Differ. 2024. PMID: 38383887 Free PMC article.
-
Kinetic discrimination of self/non-self RNA by the ATPase activity of RIG-I and MDA5.BMC Biol. 2015 Jul 28;13:54. doi: 10.1186/s12915-015-0166-9. BMC Biol. 2015. PMID: 26215161 Free PMC article.
-
Function conservation and disparities of zebrafish and human LGP2 genes in fish and mammalian cells responsive to poly(I:C).Front Immunol. 2022 Aug 17;13:985792. doi: 10.3389/fimmu.2022.985792. eCollection 2022. Front Immunol. 2022. PMID: 36059486 Free PMC article.
References
-
- Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell. 124:783–801. - PubMed
-
- Beutler, B., C. Eidenschenk, K. Crozat, J.L. Imler, O. Takeuchi, J.A. Hoffmann, and S. Akira. 2007. Genetic analysis of resistance to viral infection. Nat. Rev. Immunol. 7:753–766. - PubMed
-
- Medzhitov, R. 2007. Recognition of microorganisms and activation of the immune response. Nature. 449:819–826. - PubMed
-
- Fujita, T., K. Onoguchi, K. Onomoto, R. Hirai, and M. Yoneyama. 2007. Triggering antiviral response by RIG-I-related RNA helicases. Biochimie. 89:754–760. - PubMed
-
- Alexopoulou, L., A.C. Holt, R. Medzhitov, and R.A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 413:732–738. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

