Glucose activates ChREBP by increasing its rate of nuclear entry and relieving repression of its transcriptional activity
- PMID: 18591247
- PMCID: PMC2527113
- DOI: 10.1074/jbc.M801539200
Glucose activates ChREBP by increasing its rate of nuclear entry and relieving repression of its transcriptional activity
Abstract
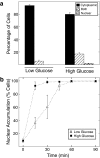
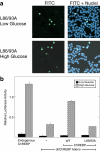
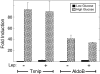
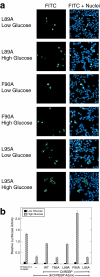
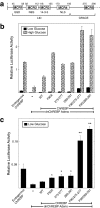
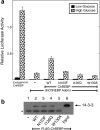
Carbohydrate response element-binding protein (ChREBP) is a glucose-responsive transcription factor that activates genes involved in de novo lipogenesis in mammals. The current model for glucose activation of ChREBP proposes that increased glucose metabolism triggers a cytoplasmic to nuclear translocation of ChREBP that is critical for activation. However, we find that ChREBP actively shuttles between the cytoplasm and nucleus in both low and high glucose in the glucose-sensitive beta cell-derived line, 832/13. Glucose stimulates a 3-fold increase in the rate of ChREBP nuclear entry, but trapping ChREBP in the nucleus by mutagenesis or with a nuclear export inhibitor does not lead to constitutive activation. In fact, mutational studies targeting the nuclear export signal of ChREBP also identified a distinct function essential for glucose-dependent transcriptional activation. From this, we conclude that an additional event independent of nuclear translocation is required for activation. The N-terminal segment of ChREBP (amino acids 1-298) has previously been shown to repress activity under basal conditions. This segment has five highly conserved regions, Mondo conserved regions 1-5 (MCR1 to -5). Based on activating mutations in MCR2 and MCR5, we propose that these two regions act coordinately to repress ChREBP in low glucose. In addition, other mutations in MCR2 and mutations in MCR3 were found to prevent glucose activation. Hence, we conclude that both relief of repression and adoption of an activating form are required for ChREBP activation.
Figures

Similar articles
-
ChREBP mediates glucose repression of peroxisome proliferator-activated receptor alpha expression in pancreatic beta-cells.J Biol Chem. 2011 Apr 15;286(15):13214-25. doi: 10.1074/jbc.M110.215467. Epub 2011 Jan 31. J Biol Chem. 2011. PMID: 21282101 Free PMC article.
-
Activation and repression of glucose-stimulated ChREBP requires the concerted action of multiple domains within the MondoA conserved region.Am J Physiol Endocrinol Metab. 2010 Oct;299(4):E665-74. doi: 10.1152/ajpendo.00349.2010. Epub 2010 Aug 3. Am J Physiol Endocrinol Metab. 2010. PMID: 20682844 Free PMC article.
-
Flightless I homolog negatively regulates ChREBP activity in cancer cells.Int J Biochem Cell Biol. 2013 Nov;45(11):2688-97. doi: 10.1016/j.biocel.2013.09.004. Epub 2013 Sep 17. Int J Biochem Cell Biol. 2013. PMID: 24055811
-
Roles of Ca2+ ions in the control of ChREBP nuclear translocation.J Endocrinol. 2012 May;213(2):115-22. doi: 10.1530/JOE-11-0480. Epub 2012 Mar 8. J Endocrinol. 2012. PMID: 22402852 Review.
-
Cross-regulation of hepatic glucose metabolism via ChREBP and nuclear receptors.Biochim Biophys Acta. 2011 Aug;1812(8):995-1006. doi: 10.1016/j.bbadis.2011.03.015. Epub 2011 Mar 29. Biochim Biophys Acta. 2011. PMID: 21453770 Review.
Cited by
-
Molecular pathways of nonalcoholic fatty liver disease development and progression.Cell Mol Life Sci. 2019 Jan;76(1):99-128. doi: 10.1007/s00018-018-2947-0. Epub 2018 Oct 20. Cell Mol Life Sci. 2019. PMID: 30343320 Free PMC article. Review.
-
Carbohydrate Sensing Through the Transcription Factor ChREBP.Front Genet. 2019 Jun 4;10:472. doi: 10.3389/fgene.2019.00472. eCollection 2019. Front Genet. 2019. PMID: 31275349 Free PMC article. Review.
-
S-Equol ameliorates insulin secretion failure through Chrebp/Txnip signaling via modulating PKA/PP2A activities.Nutr Metab (Lond). 2020 Jan 14;17:7. doi: 10.1186/s12986-020-0426-8. eCollection 2020. Nutr Metab (Lond). 2020. PMID: 31956333 Free PMC article.
-
Minireview: Thioredoxin-interacting protein: regulation and function in the pancreatic β-cell.Mol Endocrinol. 2014 Aug;28(8):1211-20. doi: 10.1210/me.2014-1095. Epub 2014 Jun 9. Mol Endocrinol. 2014. PMID: 24911120 Free PMC article. Review.
-
Partitioning of MLX-Family Transcription Factors to Lipid Droplets Regulates Metabolic Gene Expression.Mol Cell. 2020 Mar 19;77(6):1251-1264.e9. doi: 10.1016/j.molcel.2020.01.014. Epub 2020 Feb 4. Mol Cell. 2020. PMID: 32023484 Free PMC article.
References
-
- Vaulont, S., Munich, A., Decaux, J. F., and Kahn, A. (1986) J. Biol. Chem. 261 7621-7625 - PubMed
-
- Katsurada, A., Iritani, N., Fukuda, H., Matsumara, Y., Nishimoto, N., Nogushi, T., and Tanaka, T. (1990) Eur. J. Biochem. 190 427-433 - PubMed
-
- Katsurada, A., Iritani, N., Fukuda, H., Matsumura, Y., Noguchi, T., and Tanaka, T. (1989) Biochim. Biophys. Acta 1004 103-107 - PubMed
-
- Ntambi, J. (1992) J. Biol. Chem. 267 10925-10930 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

