Expression and function of Nkx6.3 in vertebrate hindbrain
- PMID: 18586225
- PMCID: PMC2555971
- DOI: 10.1016/j.brainres.2008.04.072
Expression and function of Nkx6.3 in vertebrate hindbrain
Abstract
Homeodomain transcription factors serve important functions in organogenesis and tissue differentiation, particularly with respect to the positional identity of individual cells. The Nkx6 subfamily controls tissue differentiation in the developing central nervous system where they function as transcriptional repressor proteins. Recent work indicates that Nkx6.3 is expressed in hindbrain V2 interneurons that co-express Nkx6.1, suggesting the possibility of functional redundancy. Here, we report that Nkx6.3 expression is specific to Chx10+ V2a interneurons but not to Gata3+ V2b interneurons of the hindbrain, and that Nkx6.3 expression appears to mark cells of the prospective medullary reticular formation. Molecular analysis of Nkx6.3 null embryonic mouse hindbrain did not reveal detectable defects in progenitor markers, motor neuron or V2 interneuron sub-types. Forced expression of Nkx6.3 and Nkx6.1 promote V2 interneuron differentiation in the developing chick hindbrain. These findings indicate Nkx6.3 function is dispensable for CNS development and lead to the proposal that absence of overt defects is due to functional compensation from a related homeodomain transcription factor.
Figures
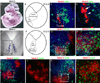
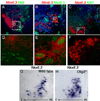
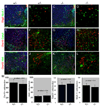
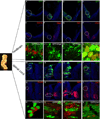
Similar articles
-
Functional dissection of the Pax6 paired domain: Roles in neural tube patterning and peripheral nervous system development.Dev Biol. 2016 May 1;413(1):86-103. doi: 10.1016/j.ydbio.2015.07.009. Epub 2015 Jul 15. Dev Biol. 2016. PMID: 26187199
-
Genetic dissection of Gata2 selective functions during specification of V2 interneurons in the developing spinal cord.Dev Neurobiol. 2015 Jul;75(7):721-37. doi: 10.1002/dneu.22244. Epub 2014 Nov 15. Dev Neurobiol. 2015. PMID: 25369423
-
Hindbrain rhombic lip is comprised of discrete progenitor cell populations allocated by Pax6.Neuron. 2005 Dec 22;48(6):933-47. doi: 10.1016/j.neuron.2005.11.031. Neuron. 2005. PMID: 16364898
-
Glucagon gene expression in the endocrine pancreas: the role of the transcription factor Pax6 in α-cell differentiation, glucagon biosynthesis and secretion.Diabetes Obes Metab. 2011 Oct;13 Suppl 1:31-8. doi: 10.1111/j.1463-1326.2011.01445.x. Diabetes Obes Metab. 2011. PMID: 21824254 Review.
-
Role of a transcription factor Pax6 in the developing vertebrate olfactory system.Dev Growth Differ. 2007 Dec;49(9):683-90. doi: 10.1111/j.1440-169X.2007.00965.x. Epub 2007 Oct 1. Dev Growth Differ. 2007. PMID: 17908181 Review.
Cited by
-
The obesity-associated gene Negr1 regulates aspects of energy balance in rat hypothalamic areas.Physiol Rep. 2014 Jul 30;2(7):e12083. doi: 10.14814/phy2.12083. Print 2014 Jul 1. Physiol Rep. 2014. PMID: 25077509 Free PMC article.
-
Deregulated NKL Homeobox Genes in B-Cell Lymphoma.Cancers (Basel). 2019 Nov 26;11(12):1874. doi: 10.3390/cancers11121874. Cancers (Basel). 2019. PMID: 31779217 Free PMC article. Review.
-
Impact of NEGR1 genetic variability on psychological traits of patients with eating disorders.Pharmacogenomics J. 2015 Jun;15(3):278-83. doi: 10.1038/tpj.2014.53. Epub 2014 Sep 23. Pharmacogenomics J. 2015. PMID: 25245582
-
Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity.Nat Genet. 2013 May;45(5):513-7. doi: 10.1038/ng.2607. Epub 2013 Apr 7. Nat Genet. 2013. PMID: 23563609 Free PMC article.
-
Molecular Organization and Patterning of the Medulla Oblongata in Health and Disease.Int J Mol Sci. 2022 Aug 17;23(16):9260. doi: 10.3390/ijms23169260. Int J Mol Sci. 2022. PMID: 36012524 Free PMC article. Review.
References
-
- Alanentalo T, Chatonnet F, Karlen M, Sulniute R, Ericson J, Andersson E, Ahlgren U. Cloning and analysis of Nkx6.3 during CNS and gastrointestinal development. Gene Expr Patterns. 2006;6:162–170. - PubMed
-
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. - PubMed
-
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45:41–53. - PubMed
-
- Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8:1510–1515. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

