Rapid and efficient construction of markerless deletions in the Escherichia coli genome
- PMID: 18567910
- PMCID: PMC2504295
- DOI: 10.1093/nar/gkn359
Rapid and efficient construction of markerless deletions in the Escherichia coli genome
Abstract
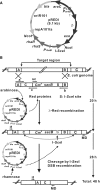
We have developed an improved and rapid genomic engineering procedure for the construction of custom-designed microorganisms. This method, which can be performed in 2 days, permits restructuring of the Escherichia coli genome via markerless deletion of selected genomic regions. The deletion process was mediated by a special plasmid, pREDI, which carries two independent inducible promoters: (i) an arabinose-inducible promoter that drives expression of lambda-Red recombination proteins, which carry out the replacement of a target genomic region with a marker-containing linear DNA cassette, and (ii) a rhamnose-inducible promoter that drives expression of I-SceI endonuclease, which stimulates deletion of the introduced marker by double-strand breakage-mediated intramolecular recombination. This genomic deletion was performed successively with only one plasmid, pREDI, simply by changing the carbon source in the bacterial growth medium from arabinose to rhamnose. The efficiencies of targeted region replacement and deletion of the inserted linear DNA cassette were nearly 70 and 100%, respectively. This rapid and efficient procedure can be adapted for use in generating a variety of genome modifications.
Figures



Similar articles
-
Scarless chromosomal gene knockout methods.Methods Mol Biol. 2011;765:43-54. doi: 10.1007/978-1-61779-197-0_3. Methods Mol Biol. 2011. PMID: 21815085
-
Fast and antibiotic free genome integration into Escherichia coli chromosome.Sci Rep. 2020 Oct 5;10(1):16510. doi: 10.1038/s41598-020-73348-x. Sci Rep. 2020. PMID: 33020519 Free PMC article.
-
An electroporation-free method based on Red recombineering for markerless deletion and genomic replacement in the Escherichia coli DH1 genome.PLoS One. 2017 Oct 24;12(10):e0186891. doi: 10.1371/journal.pone.0186891. eCollection 2017. PLoS One. 2017. PMID: 29065183 Free PMC article.
-
[Markerless DNA deletion based on Red recombination and in vivo I-Sec I endonuclease cleavage in Escherichia coli chromosome].Sheng Wu Gong Cheng Xue Bao. 2016 Jan;32(1):114-26. Sheng Wu Gong Cheng Xue Bao. 2016. PMID: 27363204 Chinese.
-
Gene replacement techniques for Escherichia coli genome modification.Folia Microbiol (Praha). 2011 May;56(3):253-63. doi: 10.1007/s12223-011-0035-z. Epub 2011 May 26. Folia Microbiol (Praha). 2011. PMID: 21614539 Review.
Cited by
-
Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system.Appl Environ Microbiol. 2015 Apr;81(7):2506-14. doi: 10.1128/AEM.04023-14. Epub 2015 Jan 30. Appl Environ Microbiol. 2015. PMID: 25636838 Free PMC article.
-
Restriction-deficient mutants and marker-less genomic modification for metabolic engineering of the solvent producer Clostridium saccharobutylicum.Biotechnol Biofuels. 2018 Sep 27;11:264. doi: 10.1186/s13068-018-1260-3. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 30275904 Free PMC article.
-
Basal transcription profiles of the rhamnose-inducible promoter PLRA3 and the development of efficient PLRA3-based systems for markerless gene deletion and a mutant library in Pichia pastoris.Curr Genet. 2019 Jun;65(3):785-798. doi: 10.1007/s00294-019-00934-6. Epub 2019 Jan 24. Curr Genet. 2019. PMID: 30680438
-
Tough nuts to crack: site-directed mutagenesis of bifidobacteria remains a challenge.Bioengineered. 2013 Jul-Aug;4(4):197-202. doi: 10.4161/bioe.23381. Epub 2013 Jan 11. Bioengineered. 2013. PMID: 23314785 Free PMC article.
-
Strategies for cloning and manipulating natural and synthetic chromosomes.Chromosome Res. 2015 Feb;23(1):57-68. doi: 10.1007/s10577-014-9455-3. Chromosome Res. 2015. PMID: 25596826 Review.
References
-
- Yu BJ, Sung BH, Koob MD, Lee CH, Lee JH, Lee WS, Kim MS, Kim SC. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat. Biotechnol. 2002;20:1018–1023. - PubMed
-
- Westers H, Dorenbos R, van Diji JM, Kabel J, Flanagan T, Devine KM, Jude F, Seror SJ, Beekman AC, Darmon E, et al. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol. Biol. Evol. 2003;20:2076–2090. - PubMed
-
- Fukiya S, Mizoguchi H, Mori H. An improved method for deleting large regions of Escherichia coli K-12 chromosome using a combination of Cre/loxP and λ Red. FEMS Microbiol. Lett. 2004;234:325–331. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

