Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes
- PMID: 18539749
- PMCID: PMC2483374
- DOI: 10.1105/tpc.108.060020
Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes
Abstract
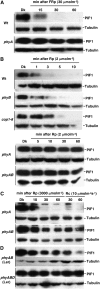
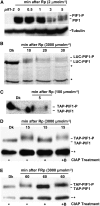
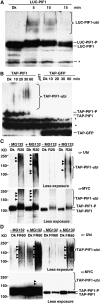
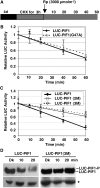
The phytochrome (phy) family of photoreceptors regulates changes in gene expression in response to red/far-red light signals in part by physically interacting with constitutively nucleus-localized phy-interacting basic helix-loop-helix transcription factors (PIFs). Here, we show that PIF1, the member with the highest affinity for phys, is strongly sensitive to the quality and quantity of light. phyA plays a dominant role in regulating the degradation of PIF1 following initial light exposure, while phyB and phyD and possibly other phys also influence PIF1 degradation after prolonged illumination. PIF1 is rapidly phosphorylated and ubiquitinated under red and far-red light before being degraded with a half-life of approximately 1 to 2 min under red light. Although PIF1 interacts with phyB through a conserved active phyB binding motif, it interacts with phyA through a novel active phyA binding motif. phy interaction is necessary but not sufficient for the light-induced phosphorylation and degradation of PIF1. Domain-mapping studies reveal that the phy interaction, light-induced degradation, and transcriptional activation domains are located at the N-terminal 150-amino acid region of PIF1. Unlike PIF3, PIF1 does not interact with the two halves of either phyA or phyB separately. Moreover, overexpression of a light-stable truncated form of PIF1 causes constitutively photomorphogenic phenotypes in the dark. Taken together, these data suggest that removal of the negative regulators (e.g., PIFs) by light-induced proteolytic degradation might be sufficient to promote photomorphogenesis.
Figures



Similar articles
-
Blue light induces degradation of the negative regulator phytochrome interacting factor 1 to promote photomorphogenic development of Arabidopsis seedlings.Genetics. 2009 May;182(1):161-71. doi: 10.1534/genetics.108.099887. Epub 2009 Mar 2. Genetics. 2009. PMID: 19255368 Free PMC article.
-
Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation.Plant Physiol. 2007 Nov;145(3):1043-51. doi: 10.1104/pp.107.105601. Epub 2007 Sep 7. Plant Physiol. 2007. PMID: 17827270 Free PMC article.
-
The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels.Plant Cell. 2008 Feb;20(2):337-52. doi: 10.1105/tpc.107.052142. Epub 2008 Feb 5. Plant Cell. 2008. PMID: 18252845 Free PMC article.
-
Multiple kinases promote light-induced degradation of PIF1.Plant Signal Behav. 2011 Aug;6(8):1119-21. doi: 10.4161/psb.6.8.16049. Epub 2011 Aug 1. Plant Signal Behav. 2011. PMID: 21758014 Free PMC article. Review.
-
Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks.Trends Plant Sci. 2007 Nov;12(11):514-521. doi: 10.1016/j.tplants.2007.10.001. Epub 2007 Oct 22. Trends Plant Sci. 2007. PMID: 17933576 Review.
Cited by
-
PHYTOCHROME INTERACTING FACTOR8 Inhibits Phytochrome A-Mediated Far-Red Light Responses in Arabidopsis.Plant Cell. 2020 Jan;32(1):186-205. doi: 10.1105/tpc.19.00515. Epub 2019 Nov 15. Plant Cell. 2020. PMID: 31732705 Free PMC article.
-
Transcriptional Regulation of Carotenoid Biosynthesis in Plants: So Many Regulators, So Little Consensus.Front Plant Sci. 2019 Aug 9;10:1017. doi: 10.3389/fpls.2019.01017. eCollection 2019. Front Plant Sci. 2019. PMID: 31447877 Free PMC article. Review.
-
Comparative metabolomic and transcriptomic analysis reveals a coexpression network of the carotenoid metabolism pathway in the panicle of Setaria italica.BMC Plant Biol. 2022 Mar 8;22(1):105. doi: 10.1186/s12870-022-03467-2. BMC Plant Biol. 2022. PMID: 35260077 Free PMC article.
-
KELCH F-BOX protein positively influences Arabidopsis seed germination by targeting PHYTOCHROME-INTERACTING FACTOR1.Proc Natl Acad Sci U S A. 2018 Apr 24;115(17):E4120-E4129. doi: 10.1073/pnas.1711919115. Epub 2018 Apr 9. Proc Natl Acad Sci U S A. 2018. PMID: 29632208 Free PMC article.
-
Cryptochromes Orchestrate Transcription Regulation of Diverse Blue Light Responses in Plants.Photochem Photobiol. 2017 Jan;93(1):112-127. doi: 10.1111/php.12663. Epub 2017 Jan 27. Photochem Photobiol. 2017. PMID: 27861972 Free PMC article. Review.
References
-
- Al-Sady, B., Ni, W., Kircher, S., Schafer, E., and Quail, P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23 439–446. - PubMed
-
- Bauer, D., Viczian, A., Kircher, S., Nobis, T., Nitschke, R., Kunkel, T., Panigrahi, K.C., Adam, E., Fejes, E., Schafer, E., and Nagy, F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16 1433–1445. - PMC - PubMed
-
- Castillon, A., Shen, H., and Huq, E. (2007). Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12 514–521. - PubMed
-
- Chen, M., Chory, J., and Fankhauser, C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38 87–117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

