Synaptic vesicle protein 2 binds adenine nucleotides
- PMID: 18524768
- PMCID: PMC2475693
- DOI: 10.1074/jbc.M800738200
Synaptic vesicle protein 2 binds adenine nucleotides
Erratum in
- J Biol Chem. 2008 Sep 5;283(36):25103
Abstract
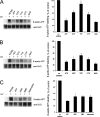
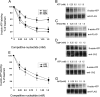
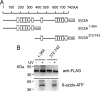
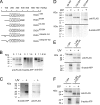
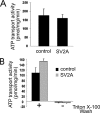
Synaptic vesicle protein 2 (SV2) is required for normal calcium-regulated secretion of hormones and neurotransmitters. Neurons lacking the two most widely expressed isoforms, SV2A and SV2B, have a reduced readily releasable pool of synaptic vesicles, indicating that SV2 contributes to vesicle priming. The presence of putative ATP-binding sites in SV2 suggested that SV2 might be an ATP-binding protein. To explore this, we examined the binding of the photoaffinity reagent 8-azido-ATP[gamma] biotin to purified, recombinant SV2 in the presence and absence of other nucleotides. Our results indicate that SV2A and SV2B bind nucleotides, with the highest affinity for adenine-containing nucleotides. SV2A contains two binding sites located in the cytoplasmic domains preceding the first and seventh transmembrane domains. These results suggest that SV2-mediated vesicle priming could be regulated by adenine nucleotides, which might provide a link between cellular energy levels and regulated secretion.
Figures

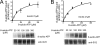

Similar articles
-
SVOP is a nucleotide binding protein.PLoS One. 2009;4(4):e5315. doi: 10.1371/journal.pone.0005315. Epub 2009 Apr 24. PLoS One. 2009. PMID: 19390693 Free PMC article.
-
Differential expression of synaptic vesicle protein 2 (SV2) isoforms.J Neurosci. 1994 Sep;14(9):5223-35. doi: 10.1523/JNEUROSCI.14-09-05223.1994. J Neurosci. 1994. PMID: 8083732 Free PMC article.
-
Cotrafficking of SV2 and synaptotagmin at the synapse.J Neurosci. 2010 Apr 21;30(16):5569-78. doi: 10.1523/JNEUROSCI.4781-09.2010. J Neurosci. 2010. PMID: 20410110 Free PMC article.
-
The Synaptic Vesicle Glycoprotein 2: Structure, Function, and Disease Relevance.ACS Chem Neurosci. 2019 Sep 18;10(9):3927-3938. doi: 10.1021/acschemneuro.9b00351. Epub 2019 Aug 23. ACS Chem Neurosci. 2019. PMID: 31394034 Free PMC article. Review.
-
KATP channel interaction with adenine nucleotides.J Mol Cell Cardiol. 2005 Jun;38(6):907-16. doi: 10.1016/j.yjmcc.2004.11.021. Epub 2005 Feb 5. J Mol Cell Cardiol. 2005. PMID: 15910875 Review.
Cited by
-
Modulation of the conformational state of the SV2A protein by an allosteric mechanism as evidenced by ligand binding assays.Br J Pharmacol. 2013 Jul;169(5):1091-101. doi: 10.1111/bph.12192. Br J Pharmacol. 2013. PMID: 23530581 Free PMC article.
-
Central presynaptic terminals are enriched in ATP but the majority lack mitochondria.PLoS One. 2015 Apr 30;10(4):e0125185. doi: 10.1371/journal.pone.0125185. eCollection 2015. PLoS One. 2015. PMID: 25928229 Free PMC article.
-
The purinergic neurotransmitter revisited: a single substance or multiple players?Pharmacol Ther. 2014 Nov;144(2):162-91. doi: 10.1016/j.pharmthera.2014.05.012. Epub 2014 Jun 2. Pharmacol Ther. 2014. PMID: 24887688 Free PMC article. Review.
-
Glucose Availability Alters Gene and Protein Expression of Several Newly Classified and Putative Solute Carriers in Mice Cortex Cell Culture and D. melanogaster.Front Cell Dev Biol. 2020 Jul 7;8:579. doi: 10.3389/fcell.2020.00579. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32733888 Free PMC article.
-
All-optical monitoring of excitation-secretion coupling demonstrates that SV2A functions downstream of evoked Ca2+ entry.J Physiol. 2022 Feb;600(3):645-654. doi: 10.1113/JP282601. Epub 2022 Jan 13. J Physiol. 2022. PMID: 34957569 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

