Introduction of an agent-based multi-scale modular architecture for dynamic knowledge representation of acute inflammation
- PMID: 18505587
- PMCID: PMC2442588
- DOI: 10.1186/1742-4682-5-11
Introduction of an agent-based multi-scale modular architecture for dynamic knowledge representation of acute inflammation
Abstract
Background: One of the greatest challenges facing biomedical research is the integration and sharing of vast amounts of information, not only for individual researchers, but also for the community at large. Agent Based Modeling (ABM) can provide a means of addressing this challenge via a unifying translational architecture for dynamic knowledge representation. This paper presents a series of linked ABMs representing multiple levels of biological organization. They are intended to translate the knowledge derived from in vitro models of acute inflammation to clinically relevant phenomenon such as multiple organ failure.
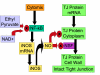
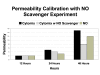
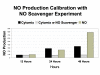
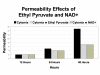
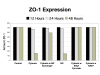
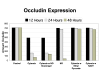
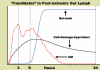
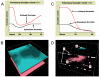
Results and discussion: ABM development followed a sequence starting with relatively direct translation from in-vitro derived rules into a cell-as-agent level ABM, leading on to concatenated ABMs into multi-tissue models, eventually resulting in topologically linked aggregate multi-tissue ABMs modeling organ-organ crosstalk. As an underlying design principle organs were considered to be functionally composed of an epithelial surface, which determined organ integrity, and an endothelial/blood interface, representing the reaction surface for the initiation and propagation of inflammation. The development of the epithelial ABM derived from an in-vitro model of gut epithelial permeability is described. Next, the epithelial ABM was concatenated with the endothelial/inflammatory cell ABM to produce an organ model of the gut. This model was validated against in-vivo models of the inflammatory response of the gut to ischemia. Finally, the gut ABM was linked to a similarly constructed pulmonary ABM to simulate the gut-pulmonary axis in the pathogenesis of multiple organ failure. The behavior of this model was validated against in-vivo and clinical observations on the cross-talk between these two organ systems.
Conclusion: A series of ABMs are presented extending from the level of intracellular mechanism to clinically observed behavior in the intensive care setting. The ABMs all utilize cell-level agents that encapsulate specific mechanistic knowledge extracted from in vitro experiments. The execution of the ABMs results in a dynamic representation of the multi-scale conceptual models derived from those experiments. These models represent a qualitative means of integrating basic scientific information on acute inflammation in a multi-scale, modular architecture as a means of conceptual model verification that can potentially be used to concatenate, communicate and advance community-wide knowledge.
Figures








Similar articles
-
Utilizing the Heterogeneity of Clinical Data for Model Refinement and Rule Discovery Through the Application of Genetic Algorithms to Calibrate a High-Dimensional Agent-Based Model of Systemic Inflammation.Front Physiol. 2021 May 19;12:662845. doi: 10.3389/fphys.2021.662845. eCollection 2021. Front Physiol. 2021. PMID: 34093225 Free PMC article.
-
Agent-based models of inflammation in translational systems biology: A decade later.Wiley Interdiscip Rev Syst Biol Med. 2019 Nov;11(6):e1460. doi: 10.1002/wsbm.1460. Epub 2019 Jul 1. Wiley Interdiscip Rev Syst Biol Med. 2019. PMID: 31260168 Free PMC article. Review.
-
Agent-based dynamic knowledge representation of Pseudomonas aeruginosa virulence activation in the stressed gut: Towards characterizing host-pathogen interactions in gut-derived sepsis.Theor Biol Med Model. 2011 Sep 19;8:33. doi: 10.1186/1742-4682-8-33. Theor Biol Med Model. 2011. PMID: 21929759 Free PMC article.
-
Introduction of a Framework for Dynamic Knowledge Representation of the Control Structure of Transplant Immunology: Employing the Power of Abstraction with a Solid Organ Transplant Agent-Based Model.Front Immunol. 2015 Nov 6;6:561. doi: 10.3389/fimmu.2015.00561. eCollection 2015. Front Immunol. 2015. PMID: 26594211 Free PMC article.
-
Addressing the translational dilemma: dynamic knowledge representation of inflammation using agent-based modeling.Crit Rev Biomed Eng. 2012;40(4):323-40. doi: 10.1615/critrevbiomedeng.v40.i4.70. Crit Rev Biomed Eng. 2012. PMID: 23140123 Review.
Cited by
-
Examining the pathogenesis of breast cancer using a novel agent-based model of mammary ductal epithelium dynamics.PLoS One. 2013 May 21;8(5):e64091. doi: 10.1371/journal.pone.0064091. Print 2013. PLoS One. 2013. PMID: 23704974 Free PMC article.
-
Agent-based model of fecal microbial transplant effect on bile acid metabolism on suppressing Clostridium difficile infection: an example of agent-based modeling of intestinal bacterial infection.J Pharmacokinet Pharmacodyn. 2014 Oct;41(5):493-507. doi: 10.1007/s10928-014-9381-1. Epub 2014 Aug 29. J Pharmacokinet Pharmacodyn. 2014. PMID: 25168489 Free PMC article.
-
A multi-scale approach reveals that NF-κB cRel enforces a B-cell decision to divide.Mol Syst Biol. 2015 Feb 13;11(1):783. doi: 10.15252/msb.20145554. Mol Syst Biol. 2015. PMID: 25680807 Free PMC article.
-
Modeling drug- and chemical-induced hepatotoxicity with systems biology approaches.Front Physiol. 2012 Dec 14;3:462. doi: 10.3389/fphys.2012.00462. eCollection 2012. Front Physiol. 2012. PMID: 23248599 Free PMC article.
-
Computational Studies of the Intestinal Host-Microbiota Interactome.Computation (Basel). 2015 Mar;3(1):2-28. doi: 10.3390/computation3010002. Epub 2015 Jan 14. Computation (Basel). 2015. PMID: 34765258 Free PMC article.
References
-
- Zhang L, Athale CA, Deisboeck TS. Development of a three-dimensional multiscale agent-based tumor model: simulating gene-protein interaction profiles, cell phenotypes and multicellular patterns in brain cancer. Journal of theoretical biology. 2007;244:96–107. doi: 10.1016/j.jtbi.2006.06.034. - DOI - PubMed
-
- An G. Agent-based computer simulation and sirs: building a bridge between basic science and clinical trials. Shock. 2001;16:266–273. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

