G protein activation by the leukotriene B4 receptor dimer. Evidence for an absence of trans-activation
- PMID: 18490452
- PMCID: PMC3258929
- DOI: 10.1074/jbc.M710419200
G protein activation by the leukotriene B4 receptor dimer. Evidence for an absence of trans-activation
Abstract
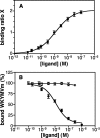
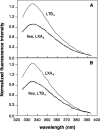
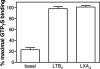
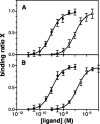
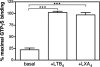
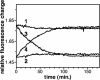
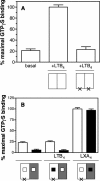
There is compelling evidence that G protein-coupled receptors exist as homo- and heterodimers, but the way these assemblies function at the molecular level remains unclear. We used here the purified leukotriene B(4) receptor BLT1 stabilized in its dimeric state to analyze how a receptor dimer activates G proteins. For this, we produced heterodimers between the wild-type BLT1 and a BLT1/ALXR chimera. The latter is no longer activated by leukotriene B(4) but is still activated by ALXR agonists. In this heterodimer, agonist binding to either one of the two protomers induced asymmetric conformational changes within the receptor dimer. Of importance, no G protein activation was observed when using a dimer where the ligand-loaded protomer was not able to trigger GDP/GTP exchange due to specific mutations in its third intracellular loop, establishing that the conformation of the agonist-free protomer is not competent for G protein activation. Taken together, these data indicate that although ligand binding to one protomer in the heterodimer is associated with cross-conformational changes, a trans-activation mechanism where the ligand-free subunit would trigger GDP/GTP exchange cannot be considered in this case for G protein activation. This observation sheds light into the way GPCR dimers, in particular heterodimers, could activate their cognate G proteins.
Figures

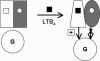
Similar articles
-
Asymmetric conformational changes in a GPCR dimer controlled by G-proteins.EMBO J. 2006 Dec 13;25(24):5693-702. doi: 10.1038/sj.emboj.7601449. Epub 2006 Nov 30. EMBO J. 2006. PMID: 17139258 Free PMC article.
-
Structure-based analysis of GPCR function: evidence for a novel pentameric assembly between the dimeric leukotriene B4 receptor BLT1 and the G-protein.J Mol Biol. 2003 Jun 13;329(4):815-29. doi: 10.1016/s0022-2836(03)00439-x. J Mol Biol. 2003. PMID: 12787680
-
Cooperative conformational changes in a G-protein-coupled receptor dimer, the leukotriene B(4) receptor BLT1.J Biol Chem. 2004 Nov 26;279(48):49664-70. doi: 10.1074/jbc.M404941200. Epub 2004 Sep 9. J Biol Chem. 2004. PMID: 15358776
-
The use of receptor G-protein fusion proteins for the study of ligand activity.Recept Channels. 2002;8(5-6):309-17. Recept Channels. 2002. PMID: 12690958 Review.
-
[Effects of newly isolated opioid peptides on G-protein activation: usefulness of [35S] GTP gamma S binding study and its practical application].Nihon Shinkei Seishin Yakurigaku Zasshi. 1998 Aug;18(4):107-16. Nihon Shinkei Seishin Yakurigaku Zasshi. 1998. PMID: 9866825 Review. Japanese.
Cited by
-
Distinct Agonist Regulation of Muscarinic Acetylcholine M2-M3 Heteromers and Their Corresponding Homomers.J Biol Chem. 2015 Jun 5;290(23):14785-96. doi: 10.1074/jbc.M115.649079. Epub 2015 Apr 27. J Biol Chem. 2015. PMID: 25918156 Free PMC article.
-
Complement component 5a receptor oligomerization and homologous receptor down-regulation.J Biol Chem. 2008 Nov 7;283(45):31038-46. doi: 10.1074/jbc.M805260200. Epub 2008 Sep 4. J Biol Chem. 2008. PMID: 18772131 Free PMC article.
-
Exploring a role for heteromerization in GPCR signalling specificity.Biochem J. 2011 Jan 1;433(1):11-8. doi: 10.1042/BJ20100458. Biochem J. 2011. PMID: 21158738 Free PMC article. Review.
-
Fluorescence correlation spectroscopy, combined with bimolecular fluorescence complementation, reveals the effects of β-arrestin complexes and endocytic targeting on the membrane mobility of neuropeptide Y receptors.Biochim Biophys Acta. 2012 Jun;1823(6):1068-81. doi: 10.1016/j.bbamcr.2012.03.002. Epub 2012 Mar 8. Biochim Biophys Acta. 2012. PMID: 22487268 Free PMC article.
-
Heterodimerization with Its splice variant blocks the ghrelin receptor 1a in a non-signaling conformation: a study with a purified heterodimer assembled into lipid discs.J Biol Chem. 2013 Aug 23;288(34):24656-65. doi: 10.1074/jbc.M113.453423. Epub 2013 Jul 9. J Biol Chem. 2013. PMID: 23839942 Free PMC article.
References
-
- Bockaert, J., Claeysen, S., Becamel, C., Pinloche, S., and Dumuis, A. (2002) Int. Rev. Cytol. 212 63-132 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

