Ribosomal RNAs are tolerant toward genetic insertions: evolutionary origin of the expansion segments
- PMID: 18456707
- PMCID: PMC2441807
- DOI: 10.1093/nar/gkn224
Ribosomal RNAs are tolerant toward genetic insertions: evolutionary origin of the expansion segments
Abstract
Ribosomal RNAs (rRNAs), assisted by ribosomal proteins, form the basic structure of the ribosome, and play critical roles in protein synthesis. Compared to prokaryotic ribosomes, eukaryotic ribosomes contain elongated rRNAs with several expansion segments and larger numbers of ribosomal proteins. To investigate architectural evolution and functional capability of rRNAs, we employed a Tn5 transposon system to develop a systematic genetic insertion of an RNA segment 31 nt in length into Escherichia coli rRNAs. From the plasmid library harboring a single rRNA operon containing random insertions, we isolated surviving clones bearing rRNAs with functional insertions that enabled rescue of the E. coli strain (Delta7rrn) in which all chromosomal rRNA operons were depleted. We identified 51 sites with functional insertions, 16 sites in 16S rRNA and 35 sites in 23S rRNA, revealing the architecture of E. coli rRNAs to be substantially flexible. Most of the insertion sites show clear tendency to coincide with the regions of the expansion segments found in eukaryotic rRNAs, implying that eukaryotic rRNAs evolved from prokaryotic rRNAs suffering genetic insertions and selections.
Figures

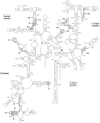


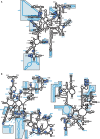
Similar articles
-
Systematic deletion of rRNAs for investigating ribosome architecture and function.Nucleic Acids Symp Ser (Oxf). 2006;(50):287-8. doi: 10.1093/nass/nrl143. Nucleic Acids Symp Ser (Oxf). 2006. PMID: 17150930
-
Correlation of the expansion segments in mammalian rRNA with the fine structure of the 80 S ribosome; a cryoelectron microscopic reconstruction of the rabbit reticulocyte ribosome at 21 A resolution.J Mol Biol. 1998 Jun 5;279(2):403-21. doi: 10.1006/jmbi.1998.1804. J Mol Biol. 1998. PMID: 9642046
-
rRNA mutations that inhibit transfer-messenger RNA activity on stalled ribosomes.J Bacteriol. 2010 Jan;192(2):553-9. doi: 10.1128/JB.01178-09. Epub 2009 Nov 6. J Bacteriol. 2010. PMID: 19897649 Free PMC article.
-
Structural and evolutionary insights into ribosomal RNA methylation.Nat Chem Biol. 2018 Feb 14;14(3):226-235. doi: 10.1038/nchembio.2569. Nat Chem Biol. 2018. PMID: 29443970 Review.
-
RNA-protein interactions in the Escherichia coli ribosome.Biochimie. 1991 Jul-Aug;73(7-8):927-36. doi: 10.1016/0300-9084(91)90134-m. Biochimie. 1991. PMID: 1720671 Review.
Cited by
-
Ribosome Subunit Stapling for Orthogonal Translation in E. coli.Angew Chem Int Ed Engl. 2015 Oct 19;54(43):12791-4. doi: 10.1002/anie.201506311. Epub 2015 Aug 26. Angew Chem Int Ed Engl. 2015. PMID: 26465656 Free PMC article.
-
Peptide Bond Formation between Aminoacyl-Minihelices by a Scaffold Derived from the Peptidyl Transferase Center.Life (Basel). 2022 Apr 12;12(4):573. doi: 10.3390/life12040573. Life (Basel). 2022. PMID: 35455064 Free PMC article.
-
Varying strength of selection contributes to the intragenomic diversity of rRNA genes.Nat Commun. 2022 Nov 25;13(1):7245. doi: 10.1038/s41467-022-34989-w. Nat Commun. 2022. PMID: 36434003 Free PMC article.
-
The GA-minor submotif as a case study of RNA modularity, prediction, and design.Wiley Interdiscip Rev RNA. 2013 Mar-Apr;4(2):181-203. doi: 10.1002/wrna.1153. Epub 2013 Feb 1. Wiley Interdiscip Rev RNA. 2013. PMID: 23378290 Free PMC article. Review.
-
Computing the origin and evolution of the ribosome from its structure - Uncovering processes of macromolecular accretion benefiting synthetic biology.Comput Struct Biotechnol J. 2015 Jul 26;13:427-47. doi: 10.1016/j.csbj.2015.07.003. eCollection 2015. Comput Struct Biotechnol J. 2015. PMID: 27096056 Free PMC article. Review.
References
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. - PubMed
-
- Wimberly BT, Brodersen DE, Clemons W.M., Jr., Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. - PubMed
-
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. - PubMed
-
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. - PubMed
-
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

