Alternative splicing regulation during C. elegans development: splicing factors as regulated targets
- PMID: 18454200
- PMCID: PMC2265522
- DOI: 10.1371/journal.pgen.1000001
Alternative splicing regulation during C. elegans development: splicing factors as regulated targets
Abstract
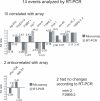
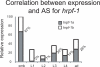
Alternative splicing generates protein diversity and allows for post-transcriptional gene regulation. Estimates suggest that 10% of the genes in Caenorhabditis elegans undergo alternative splicing. We constructed a splicing-sensitive microarray to detect alternative splicing for 352 cassette exons and tested for changes in alternative splicing of these genes during development. We found that the microarray data predicted that 62/352 (approximately 18%) of the alternative splicing events studied show a strong change in the relative levels of the spliced isoforms (>4-fold) during development. Confirmation of the microarray data by RT-PCR was obtained for 70% of randomly selected genes tested. Among the genes with the most developmentally regulated alternatively splicing was the hnRNP F/H splicing factor homolog, W02D3.11 - now named hrpf-1. For the cassette exon of hrpf-1, the inclusion isoform comprises 65% of hrpf-1 steady state messages in embryos but only 0.1% in the first larval stage. This dramatic change in the alternative splicing of an alternative splicing factor suggests a complex cascade of splicing regulation during development. We analyzed splicing in embryos from a strain with a mutation in the splicing factor sym-2, another hnRNP F/H homolog. We found that approximately half of the genes with large alternative splicing changes between the embryo and L1 stages are regulated by sym-2 in embryos. An analysis of the role of nonsense-mediated decay in regulating steady-state alternative mRNA isoforms was performed. We found that 8% of the 352 events studied have alternative isoforms whose relative steady-state levels in embryos change more than 4-fold in a nonsense-mediated decay mutant, including hrpf-1. Strikingly, 53% of these alternative splicing events that are affected by NMD in our experiment are not obvious substrates for NMD based on the presence of premature termination codons. This suggests that the targeting of splicing factors by NMD may have downstream effects on alternative splicing regulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Global analysis of alternative splicing uncovers developmental regulation of nonsense-mediated decay in C. elegans.RNA. 2009 Sep;15(9):1652-60. doi: 10.1261/rna.1711109. Epub 2009 Jul 17. RNA. 2009. PMID: 19617316 Free PMC article.
-
Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway.Genes Dev. 2007 May 1;21(9):1075-85. doi: 10.1101/gad.417707. Epub 2007 Apr 16. Genes Dev. 2007. PMID: 17437990 Free PMC article.
-
Alternative splicing and the steady-state ratios of mRNA isoforms generated by it are under strong stabilizing selection in Caenorhabditis elegans.Mol Biol Evol. 2008 Nov;25(11):2431-7. doi: 10.1093/molbev/msn181. Epub 2008 Aug 20. Mol Biol Evol. 2008. PMID: 18718918 Free PMC article.
-
Pre-mRNA splicing and its regulation in Caenorhabditis elegans.WormBook. 2012 Mar 21:1-21. doi: 10.1895/wormbook.1.31.2. WormBook. 2012. PMID: 22467343 Free PMC article. Review.
-
Alternative splicing in C. elegans.WormBook. 2005 Sep 26:1-13. doi: 10.1895/wormbook.1.31.1. WormBook. 2005. PMID: 18050427 Free PMC article. Review.
Cited by
-
Changes in gene expression associated with reproductive maturation in wild female baboons.Genome Biol Evol. 2012;4(2):102-9. doi: 10.1093/gbe/evr134. Epub 2011 Dec 8. Genome Biol Evol. 2012. PMID: 22155733 Free PMC article.
-
Two Forms of Sexual Dimorphism in Gene Expression in Drosophila melanogaster: Their Coincidence and Evolutionary Genetics.Mol Biol Evol. 2023 May 2;40(5):msad091. doi: 10.1093/molbev/msad091. Mol Biol Evol. 2023. PMID: 37116199 Free PMC article.
-
Identification of global alternative splicing and sex-specific splicing via comparative transcriptome analysis of gonads of Chinese tongue sole ( Cynoglossus semilaevis).Zool Res. 2022 May 18;43(3):319-330. doi: 10.24272/j.issn.2095-8137.2021.406. Zool Res. 2022. PMID: 35301828 Free PMC article.
-
Antagonistic factors control the unproductive splicing of SC35 terminal intron.Nucleic Acids Res. 2010 Mar;38(4):1353-66. doi: 10.1093/nar/gkp1086. Epub 2009 Dec 3. Nucleic Acids Res. 2010. PMID: 19965769 Free PMC article.
-
Nutritional control of mRNA isoform expression during developmental arrest and recovery in C. elegans.Genome Res. 2012 Oct;22(10):1920-9. doi: 10.1101/gr.133587.111. Epub 2012 Apr 26. Genome Res. 2012. PMID: 22539650 Free PMC article.
References
-
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. - PubMed
-
- Pick M, Flores-Flores C, Soreq H. From brain to blood: alternative splicing evidence for the cholinergic basis of Mammalian stress responses. Ann N Y Acad Sci. 2004;1018:85–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

