Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency
- PMID: 18423197
- PMCID: PMC2615249
- DOI: 10.1016/j.cell.2008.03.028
Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency
Erratum in
- Cell. 2008 Jul 25;134(2):365
Abstract
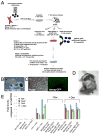
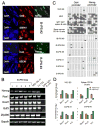
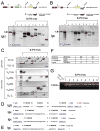
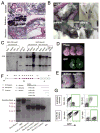
Pluripotent cells can be derived from fibroblasts by ectopic expression of defined transcription factors. A fundamental unresolved question is whether terminally differentiated cells can be reprogrammed to pluripotency. We utilized transgenic and inducible expression of four transcription factors (Oct4, Sox2, Klf4, and c-Myc) to reprogram mouse B lymphocytes. These factors were sufficient to convert nonterminally differentiated B cells to a pluripotent state. However, reprogramming of mature B cells required additional interruption with the transcriptional state maintaining B cell identity by either ectopic expression of the myeloid transcription factor CCAAT/enhancer-binding-protein-alpha (C/EBPalpha) or specific knockdown of the B cell transcription factor Pax5. Multiple iPS lines were clonally derived from both nonfully and fully differentiated B lymphocytes, which gave rise to adult chimeras with germline contribution, and to late-term embryos when injected into tetraploid blastocysts. Our study provides definite proof for the direct nuclear reprogramming of terminally differentiated adult cells to pluripotency.
Figures



Similar articles
-
Heterokaryon-based reprogramming of human B lymphocytes for pluripotency requires Oct4 but not Sox2.PLoS Genet. 2008 Sep 5;4(9):e1000170. doi: 10.1371/journal.pgen.1000170. PLoS Genet. 2008. PMID: 18773085 Free PMC article.
-
Reprogramming of pancreatic beta cells into induced pluripotent stem cells.Curr Biol. 2008 Jun 24;18(12):890-4. doi: 10.1016/j.cub.2008.05.010. Epub 2008 May 22. Curr Biol. 2008. PMID: 18501604 Free PMC article.
-
Attempting to Convert Primed Porcine Embryonic Stem Cells into a Naive State Through the Overexpression of Reprogramming Factors.Cell Reprogram. 2018 Oct;20(5):289-300. doi: 10.1089/cell.2017.0071. Cell Reprogram. 2018. PMID: 30277824
-
Exploring the reprogramming potential of B cells and comprehending its clinical and therapeutic perspective.Transpl Immunol. 2023 Jun;78:101804. doi: 10.1016/j.trim.2023.101804. Epub 2023 Mar 13. Transpl Immunol. 2023. PMID: 36921730 Review.
-
Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues.Cell. 2010 Nov 12;143(4):508-25. doi: 10.1016/j.cell.2010.10.008. Cell. 2010. PMID: 21074044 Free PMC article. Review.
Cited by
-
AP-1 is a temporally regulated dual gatekeeper of reprogramming to pluripotency.Proc Natl Acad Sci U S A. 2021 Jun 8;118(23):e2104841118. doi: 10.1073/pnas.2104841118. Proc Natl Acad Sci U S A. 2021. PMID: 34088849 Free PMC article.
-
Establishment and optimal culture conditions of microrna-induced pluripotent stem cells generated from HEK293 cells via transfection of microrna-302s expression vector.Nagoya J Med Sci. 2012 Feb;74(1-2):157-65. Nagoya J Med Sci. 2012. PMID: 22515122 Free PMC article.
-
Excessive Cellular Proliferation Negatively Impacts Reprogramming Efficiency of Human Fibroblasts.Stem Cells Transl Med. 2015 Oct;4(10):1101-8. doi: 10.5966/sctm.2014-0217. Epub 2015 Aug 7. Stem Cells Transl Med. 2015. PMID: 26253715 Free PMC article.
-
Induced pluripotent stem cells: applications in regenerative medicine, disease modeling, and drug discovery.Front Cell Dev Biol. 2015 Feb 2;3:2. doi: 10.3389/fcell.2015.00002. eCollection 2015. Front Cell Dev Biol. 2015. PMID: 25699255 Free PMC article. Review.
-
Resetting epigenetic signatures to induce somatic cell reprogramming.Cell Mol Life Sci. 2013 Apr;70(8):1413-24. doi: 10.1007/s00018-012-1137-8. Epub 2012 Aug 30. Cell Mol Life Sci. 2013. PMID: 22932957 Free PMC article. Review.
References
-
- Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981;27:381–390. - PubMed
-
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science (Science Express) 2008 - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

