Genetic analyses of the role of RCE1 in RAS membrane association and transformation
- PMID: 18413262
- PMCID: PMC2386417
- DOI: 10.1016/S0076-6879(07)38026-9
Genetic analyses of the role of RCE1 in RAS membrane association and transformation
Abstract
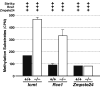
Proteins terminating with a CAAX motif, such as the nuclear lamins and the RAS family of proteins, undergo post-translational modification of a carboxyl-terminal cysteine with an isoprenyl lipid--a process called protein prenylation. After prenylation, the last three residues of CAAX proteins are clipped off by an endoprotease of the endoplasmic reticulum. RCE1 is responsible for the endoproteolytic processing of the RAS proteins and is likely responsible for endoproteolytic processing of the vast majority of CAAX proteins. Prenylation has been shown to be essential for the proper intracellular targeting and function of several CAAX proteins, but the physiologic importance of the endoprotease step has remained less certain. Here, we will review methods that have been used to define the physiologic importance of the endoproteolytic processing step of CAAX protein processing.
Figures



Similar articles
-
On the physiological importance of endoproteolysis of CAAX proteins: heart-specific RCE1 knockout mice develop a lethal cardiomyopathy.J Biol Chem. 2004 Feb 6;279(6):4729-36. doi: 10.1074/jbc.M310081200. Epub 2003 Nov 18. J Biol Chem. 2004. PMID: 14625273
-
Absence of the CAAX endoprotease Rce1: effects on cell growth and transformation.Mol Cell Biol. 2002 Jan;22(1):171-81. doi: 10.1128/MCB.22.1.171-181.2002. Mol Cell Biol. 2002. PMID: 11739732 Free PMC article.
-
Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases.Mol Biol Cell. 2005 Apr;16(4):1606-16. doi: 10.1091/mbc.e04-11-0960. Epub 2005 Jan 19. Mol Biol Cell. 2005. PMID: 15659645 Free PMC article.
-
Rce1: mechanism and inhibition.Crit Rev Biochem Mol Biol. 2018 Apr;53(2):157-174. doi: 10.1080/10409238.2018.1431606. Epub 2018 Feb 9. Crit Rev Biochem Mol Biol. 2018. PMID: 29424242 Free PMC article. Review.
-
Post-prenylation-processing enzymes as new targets in oncogenesis.Nat Rev Cancer. 2005 May;5(5):405-12. doi: 10.1038/nrc1612. Nat Rev Cancer. 2005. PMID: 15864282 Review.
Cited by
-
Photoaffinity labeling of Ras converting enzyme using peptide substrates that incorporate benzoylphenylalanine (Bpa) residues: improved labeling and structural implications.Bioorg Med Chem. 2011 Dec 15;19(24):7559-69. doi: 10.1016/j.bmc.2011.10.027. Epub 2011 Oct 18. Bioorg Med Chem. 2011. PMID: 22079863 Free PMC article.
-
Evaluation of a cell penetrating prenylated peptide lacking an intrinsic fluorophore via in situ click reaction.Bioorg Med Chem Lett. 2011 Sep 1;21(17):4998-5001. doi: 10.1016/j.bmcl.2011.04.138. Epub 2011 May 6. Bioorg Med Chem Lett. 2011. PMID: 21632248 Free PMC article.
-
Targeting Mitogen-Activated Protein Kinase Signaling in Mouse Models of Cardiomyopathy Caused by Lamin A/C Gene Mutations.Methods Enzymol. 2016;568:557-80. doi: 10.1016/bs.mie.2015.07.028. Epub 2015 Oct 24. Methods Enzymol. 2016. PMID: 26795484 Free PMC article.
-
Post-translational modification of KRAS: potential targets for cancer therapy.Acta Pharmacol Sin. 2021 Aug;42(8):1201-1211. doi: 10.1038/s41401-020-00542-y. Epub 2020 Oct 21. Acta Pharmacol Sin. 2021. PMID: 33087838 Free PMC article. Review.
-
YcaO-Dependent Posttranslational Amide Activation: Biosynthesis, Structure, and Function.Chem Rev. 2017 Apr 26;117(8):5389-5456. doi: 10.1021/acs.chemrev.6b00623. Epub 2017 Mar 3. Chem Rev. 2017. PMID: 28256131 Free PMC article. Review.
References
-
- Aiyagari AL, Taylor BR, Aurora V, Young SG, Shannon KM. Hematologic effects of inactivating the Ras processing enzyme Rce1. Blood. 2003;101:2250–2252. - PubMed
-
- Ashby MN. CaaX converting enzymes. Curr Opin Lipidol. 1998;9:99–102. - PubMed
-
- Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Young SG. Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J Biol Chem. 2000;275:17605–17610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

