Domain stabilities in protein kinase R (PKR): evidence for weak interdomain interactions
- PMID: 18393532
- PMCID: PMC2729556
- DOI: 10.1021/bi702211j
Domain stabilities in protein kinase R (PKR): evidence for weak interdomain interactions
Abstract
PKR (protein kinase R) is induced by interferon and is a key component of the innate immunity antiviral pathway. Upon binding dsRNA, PKR undergoes autophosphorylation reactions that activate the kinase, leading it to phosphorylate eIF2alpha, thus inhibiting protein synthesis in virally infected cells. PKR contains a dsRNA-binding domain (dsRBD) and a kinase domain. The dsRBD is composed of two tandem dsRNA-binding motifs. An autoinhibition model for PKR has been proposed, whereby dsRNA binding activates the enzyme by inducing a conformational change that relieves the latent enzyme of the inhibition that is mediated by the interaction of the dsRBD with the kinase. However, recent biophysical data support an open conformation for the latent enzyme, where activation is mediated by dimerization of PKR induced upon binding dsRNA. We have probed the importance of interdomain contacts by comparing the relative stabilities of isolated domains with the same domain in the context of the intact enzyme using equilibrium chemical denaturation experiments. The two dsRNA-binding motifs fold independently, with the C-terminal motif exhibiting greater stability. The kinase domain is stabilized by about 1.5 kcal/mol in the context of the holenzyme, and we detect low-affinity binding of the kinase and dsRBD constructs in solution, indicating that these domains interact weakly. Limited proteolysis measurements confirm the expected domain boundaries and reveal that the activation loop in the kinase is accessible to cleavage and unstructured. Autophosphorylation induces a conformation change that blocks proteolysis of the activation loop.
Figures

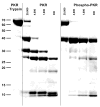
 ) and dsRBD (
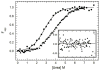
) and dsRBD ( ). and the lines are the respective two- and three-state fits. The inset shows the residuals. The best-fit parameters are in Table 1.
). and the lines are the respective two- and three-state fits. The inset shows the residuals. The best-fit parameters are in Table 1.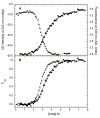
 ) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm (
) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm ( ). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.
). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.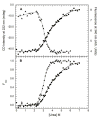
 ) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm (
) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm ( ). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.
). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.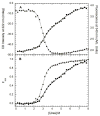
 ) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm (
) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm ( ). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.
). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.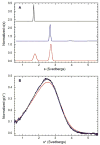
Similar articles
-
Unactivated PKR exists in an open conformation capable of binding nucleotides.Biochemistry. 2006 Aug 1;45(30):9074-84. doi: 10.1021/bi060567d. Biochemistry. 2006. PMID: 16866353 Free PMC article.
-
trans-Autophosphorylation by the isolated kinase domain is not sufficient for dimerization or activation of the dsRNA-activated protein kinase PKR.Biochemistry. 2004 Aug 31;43(34):11027-34. doi: 10.1021/bi0360105. Biochemistry. 2004. PMID: 15323561
-
Mechanism of interaction of the double-stranded RNA (dsRNA) binding domain of protein kinase R with short dsRNA sequences.Biochemistry. 2007 Jan 9;46(1):55-65. doi: 10.1021/bi061531o. Biochemistry. 2007. PMID: 17198375
-
Activation of PKR: an open and shut case?Trends Biochem Sci. 2007 Feb;32(2):57-62. doi: 10.1016/j.tibs.2006.12.003. Epub 2006 Dec 29. Trends Biochem Sci. 2007. PMID: 17196820 Free PMC article. Review.
-
Protein kinase R and the inflammasome.J Interferon Cytokine Res. 2014 Jun;34(6):447-54. doi: 10.1089/jir.2014.0008. J Interferon Cytokine Res. 2014. PMID: 24905201 Review.
Cited by
-
Improved control of tuberculosis and activation of macrophages in mice lacking protein kinase R.PLoS One. 2012;7(2):e30512. doi: 10.1371/journal.pone.0030512. Epub 2012 Feb 16. PLoS One. 2012. PMID: 22359543 Free PMC article.
-
Contribution of dsRBD2 to PKR Activation.ACS Omega. 2021 Apr 19;6(17):11367-11374. doi: 10.1021/acsomega.1c00343. eCollection 2021 May 4. ACS Omega. 2021. PMID: 34056292 Free PMC article.
-
Domain interactions in adenovirus VAI RNA mediate high-affinity PKR binding.J Mol Biol. 2014 Mar 20;426(6):1285-95. doi: 10.1016/j.jmb.2013.12.019. Epub 2014 Jan 4. J Mol Biol. 2014. PMID: 24394721 Free PMC article.
-
Monitoring activation of the antiviral pattern recognition receptors RIG-I and PKR by limited protease digestion and native PAGE.J Vis Exp. 2014 Jul 29;(89):e51415. doi: 10.3791/51415. J Vis Exp. 2014. PMID: 25146252 Free PMC article.
-
Regulation of PKR by RNA: formation of active and inactive dimers.Biochemistry. 2015 Nov 10;54(44):6663-72. doi: 10.1021/acs.biochem.5b01046. Epub 2015 Oct 26. Biochemistry. 2015. PMID: 26488609 Free PMC article.
References
-
- Clemens MJ, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. - PubMed
-
- Kaufman RJ. The double stranded RNA-activated protein kinase PKR. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. pp. 503–528.
-
- Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. - PubMed
-
- Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. - PubMed
-
- Williams BR. Signal integration via PKR. Sci STKE. 2001:RE2. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

