SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis
- PMID: 18374647
- PMCID: PMC2366111
- DOI: 10.1016/j.molcel.2008.01.013
SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis
Abstract
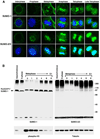
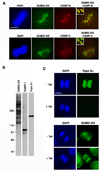
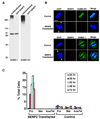
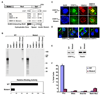
SUMOylation is essential for cell-cycle regulation in invertebrates; however, its functions during the mammalian cell cycle are largely uncharacterized. Mammals express three SUMO paralogs: SUMO-1, SUMO-2, and SUMO-3 (SUMO-2 and SUMO-3 are 96% identical and referred to as SUMO-2/3). We found that SUMO-2/3 localize to centromeres and condensed chromosomes, whereas SUMO-1 localizes to the mitotic spindle and spindle midzone, indicating that SUMO paralogs regulate distinct mitotic processes in mammalian cells. Consistent with this, global inhibition of SUMOylation caused a prometaphase arrest due to defects in targeting the microtubule motor protein CENP-E to kinetochores. CENP-E was found to be modified specifically by SUMO-2/3 and to possess SUMO-2/3 polymeric chain-binding activity essential for kinetochore localization. Our findings indicate that SUMOylation is a key regulator of the mammalian cell cycle, with SUMO-1 and SUMO-2/3 modification of different proteins regulating distinct processes.
Figures


Similar articles
-
Poly-SUMO-2/3 chain modification of Nuf2 facilitates CENP-E kinetochore localization and chromosome congression during mitosis.Cell Cycle. 2021 May;20(9):855-873. doi: 10.1080/15384101.2021.1907509. Epub 2021 Apr 28. Cell Cycle. 2021. PMID: 33910471 Free PMC article.
-
Mps1 is SUMO-modified during the cell cycle.Oncotarget. 2016 Jan 19;7(3):3158-70. doi: 10.18632/oncotarget.6552. Oncotarget. 2016. PMID: 26675261 Free PMC article.
-
SUMOylation in control of accurate chromosome segregation during mitosis.Curr Protein Pept Sci. 2012 Aug;13(5):467-81. doi: 10.2174/138920312802430563. Curr Protein Pept Sci. 2012. PMID: 22812528 Free PMC article. Review.
-
The fate of metaphase kinetochores is weighed in the balance of SUMOylation during S phase.Cell Cycle. 2010 Aug 15;9(16):3194-201. doi: 10.4161/cc.9.16.12619. Epub 2010 Aug 9. Cell Cycle. 2010. PMID: 20724819 Free PMC article. Review.
-
RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin.Mol Biol Cell. 2009 Jan;20(1):410-8. doi: 10.1091/mbc.e08-05-0511. Epub 2008 Oct 22. Mol Biol Cell. 2009. PMID: 18946085 Free PMC article.
Cited by
-
A comprehensive compilation of SUMO proteomics.Nat Rev Mol Cell Biol. 2016 Sep;17(9):581-95. doi: 10.1038/nrm.2016.81. Epub 2016 Jul 20. Nat Rev Mol Cell Biol. 2016. PMID: 27435506
-
TRAMM/TrappC12 plays a role in chromosome congression, kinetochore stability, and CENP-E recruitment.J Cell Biol. 2015 Apr 27;209(2):221-34. doi: 10.1083/jcb.201501090. J Cell Biol. 2015. PMID: 25918224 Free PMC article.
-
Emerging roles of sumoylation in the regulation of actin, microtubules, intermediate filaments, and septins.Cytoskeleton (Hoboken). 2015 Jul;72(7):305-39. doi: 10.1002/cm.21226. Epub 2015 Aug 22. Cytoskeleton (Hoboken). 2015. PMID: 26033929 Free PMC article. Review.
-
Analysis of global sumoylation changes occurring during keratinocyte differentiation.PLoS One. 2012;7(1):e30165. doi: 10.1371/journal.pone.0030165. Epub 2012 Jan 23. PLoS One. 2012. PMID: 22291911 Free PMC article.
-
A novel SUMO1-specific interacting motif in dipeptidyl peptidase 9 (DPP9) that is important for enzymatic regulation.J Biol Chem. 2012 Dec 28;287(53):44320-9. doi: 10.1074/jbc.M112.397224. Epub 2012 Nov 14. J Biol Chem. 2012. PMID: 23152501 Free PMC article.
References
-
- Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9:1169–1182. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

