Arrangement of L2 within the papillomavirus capsid
- PMID: 18367526
- PMCID: PMC2395198
- DOI: 10.1128/JVI.02726-07
Arrangement of L2 within the papillomavirus capsid
Abstract
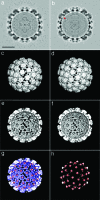
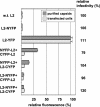
Papillomaviruses are a family of nonenveloped DNA tumor viruses. Some sexually transmitted human papillomavirus (HPV) types, including HPV type 16 (HPV16), cause cancer of the uterine cervix. Papillomaviruses encode two capsid proteins, L1 and L2. The major capsid protein, L1, can assemble spontaneously into a 72-pentamer icosahedral structure that closely resembles native virions. Although the minor capsid protein, L2, is not required for capsid formation, it is thought to participate in encapsidation of the viral genome and plays a number of essential roles in the viral infectious entry pathway. The abundance of L2 and its arrangement within the virion remain unclear. To address these questions, we developed methods for serial propagation of infectious HPV16 capsids (pseudoviruses) in cultured human cell lines. Biochemical analysis of capsid preparations produced using various methods showed that up to 72 molecules of L2 can be incorporated per capsid. Cryoelectron microscopy and image reconstruction analysis of purified capsids revealed an icosahedrally ordered L2-specific density beneath the axial lumen of each L1 capsomer. The relatively close proximity of these L2 density buttons to one another raised the possibility of homotypic L2 interactions within assembled virions. The concept that the N and C termini of neighboring L2 molecules can be closely apposed within the capsid was supported using bimolecular fluorescence complementation or "split GFP" technology. This structural information should facilitate investigation of L2 function during the assembly and entry phases of the papillomavirus life cycle.
Figures

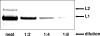

Similar articles
-
High resolution cryo EM analysis of HPV16 identifies minor structural protein L2 and describes capsid flexibility.Sci Rep. 2021 Feb 10;11(1):3498. doi: 10.1038/s41598-021-83076-5. Sci Rep. 2021. PMID: 33568731 Free PMC article.
-
Phosphorylation of Human Papillomavirus Type 16 L2 Contributes to Efficient Virus Infectious Entry.J Virol. 2019 Jun 14;93(13):e00128-19. doi: 10.1128/JVI.00128-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996086 Free PMC article.
-
The papillomavirus major capsid protein L1.Virology. 2013 Oct;445(1-2):169-74. doi: 10.1016/j.virol.2013.05.038. Epub 2013 Jun 22. Virology. 2013. PMID: 23800545 Free PMC article. Review.
-
L2, the minor capsid protein of papillomavirus.Virology. 2013 Oct;445(1-2):175-86. doi: 10.1016/j.virol.2013.04.017. Epub 2013 May 17. Virology. 2013. PMID: 23689062 Free PMC article. Review.
-
Cryoelectron Microscopy Maps of Human Papillomavirus 16 Reveal L2 Densities and Heparin Binding Site.Structure. 2017 Feb 7;25(2):253-263. doi: 10.1016/j.str.2016.12.001. Epub 2017 Jan 5. Structure. 2017. PMID: 28065506
Cited by
-
Interaction of human tumor viruses with host cell surface receptors and cell entry.Viruses. 2015 May 22;7(5):2592-617. doi: 10.3390/v7052592. Viruses. 2015. PMID: 26008702 Free PMC article. Review.
-
Identification of HPV16's E6 gene in suspected cases of cervical lesions and docking Study of its L1 protein with active components of Echinacea purpurae.Afr Health Sci. 2022 Mar;22(1):98-105. doi: 10.4314/ahs.v22i1.13. Afr Health Sci. 2022. PMID: 36032471 Free PMC article.
-
Tetraspanin CD151 mediates papillomavirus type 16 endocytosis.J Virol. 2013 Mar;87(6):3435-46. doi: 10.1128/JVI.02906-12. Epub 2013 Jan 9. J Virol. 2013. PMID: 23302890 Free PMC article.
-
Diversity in Proprotein Convertase Reactivity among Human Papillomavirus Types.Viruses. 2023 Dec 26;16(1):39. doi: 10.3390/v16010039. Viruses. 2023. PMID: 38257739 Free PMC article.
-
Targeting Endosomal Recycling Pathways by Bacterial and Viral Pathogens.Front Cell Dev Biol. 2021 Mar 4;9:648024. doi: 10.3389/fcell.2021.648024. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33748141 Free PMC article. Review.
References
-
- Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116120-130. - PubMed
-
- Buck, C. B., D. V. Pastrana, D. R. Lowy, and J. T. Schiller. 2005. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol. Med. 119445-462. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

