Orchestration of the S-phase and DNA damage checkpoint pathways by replication forks from early origins
- PMID: 18347065
- PMCID: PMC2290838
- DOI: 10.1083/jcb.200706009
Orchestration of the S-phase and DNA damage checkpoint pathways by replication forks from early origins
Abstract
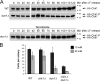
The S-phase checkpoint activated at replication forks coordinates DNA replication when forks stall because of DNA damage or low deoxyribonucleotide triphosphate pools. We explore the involvement of replication forks in coordinating the S-phase checkpoint using dun1Delta cells that have a defect in the number of stalled forks formed from early origins and are dependent on the DNA damage Chk1p pathway for survival when replication is stalled. We show that providing additional origins activated in early S phase and establishing a paused fork at a replication fork pause site restores S-phase checkpoint signaling to chk1Delta dun1Delta cells and relieves the reliance on the DNA damage checkpoint pathway. Origin licensing and activation are controlled by the cyclin-Cdk complexes. Thus, oncogene-mediated deregulation of cyclins in the early stages of cancer development could contribute to genomic instability through a deficiency in the forks required to establish the S-phase checkpoint.
Figures



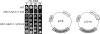



Similar articles
-
A role for Saccharomyces cerevisiae Chk1p in the response to replication blocks.Mol Biol Cell. 2004 Sep;15(9):4051-63. doi: 10.1091/mbc.e03-11-0792. Epub 2004 Jun 30. Mol Biol Cell. 2004. PMID: 15229282 Free PMC article.
-
A central role for DNA replication forks in checkpoint activation and response.Mol Cell. 2003 May;11(5):1323-36. doi: 10.1016/s1097-2765(03)00169-2. Mol Cell. 2003. PMID: 12769855
-
Genetic Evidence for Roles of Yeast Mitotic Cyclins at Single-Stranded Gaps Created by DNA Replication.G3 (Bethesda). 2018 Feb 2;8(2):737-752. doi: 10.1534/g3.117.300537. G3 (Bethesda). 2018. PMID: 29279302 Free PMC article.
-
Cyclin-dependent kinases and S phase control in mammalian cells.Cell Cycle. 2003 Jul-Aug;2(4):316-24. Cell Cycle. 2003. PMID: 12851482 Review.
-
ATR/Mec1: coordinating fork stability and repair.Curr Opin Cell Biol. 2009 Apr;21(2):237-44. doi: 10.1016/j.ceb.2009.01.017. Epub 2009 Feb 21. Curr Opin Cell Biol. 2009. PMID: 19230642 Review.
Cited by
-
Activation of the S-phase checkpoint inhibits degradation of the F-box protein Dia2.Mol Cell Biol. 2010 Jan;30(1):160-71. doi: 10.1128/MCB.00612-09. Mol Cell Biol. 2010. PMID: 19858292 Free PMC article.
-
Clb6-Cdc28 Promotes Ribonucleotide Reductase Subcellular Redistribution during S Phase.Mol Cell Biol. 2018 Feb 27;38(6):e00497-17. doi: 10.1128/MCB.00497-17. Print 2018 Mar 15. Mol Cell Biol. 2018. PMID: 29263158 Free PMC article.
-
Fragile genomic sites are associated with origins of replication.Genome Biol Evol. 2009 Sep 9;1:350-63. doi: 10.1093/gbe/evp034. Genome Biol Evol. 2009. PMID: 20333204 Free PMC article.
-
Divergent S phase checkpoint activation arising from prereplicative complex deficiency controls cell survival.Mol Biol Cell. 2009 Sep;20(17):3953-64. doi: 10.1091/mbc.e09-01-0022. Epub 2009 Jul 8. Mol Biol Cell. 2009. PMID: 19587119 Free PMC article.
-
The DNA damage response pathway contributes to the stability of chromosome III derivatives lacking efficient replicators.PLoS Genet. 2010 Dec 2;6(12):e1001227. doi: 10.1371/journal.pgen.1001227. PLoS Genet. 2010. PMID: 21151954 Free PMC article.
References
-
- Alcasabas, A.A., A.J. Osborn, J. Bachant, F. Hu, P.J. Werler, K. Bousset, K. Furuya, J.F. Diffley, A.M. Carr, and S.J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958–965. - PubMed
-
- Allen, J.B., Z. Zhou, W. Siede, E.C. Friedberg, and S.J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401–2415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

