The Arabidopsis P4-ATPase ALA3 localizes to the golgi and requires a beta-subunit to function in lipid translocation and secretory vesicle formation
- PMID: 18344284
- PMCID: PMC2329932
- DOI: 10.1105/tpc.107.054767
The Arabidopsis P4-ATPase ALA3 localizes to the golgi and requires a beta-subunit to function in lipid translocation and secretory vesicle formation
Abstract
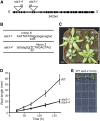
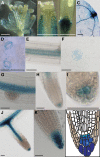
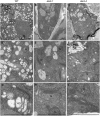
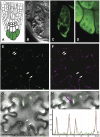
Vesicle budding in eukaryotes depends on the activity of lipid translocases (P(4)-ATPases) that have been implicated in generating lipid asymmetry between the two leaflets of the membrane and in inducing membrane curvature. We show that Aminophospholipid ATPase3 (ALA3), a member of the P(4)-ATPase subfamily in Arabidopsis thaliana, localizes to the Golgi apparatus and that mutations of ALA3 result in impaired growth of roots and shoots. The growth defect is accompanied by failure of the root cap to release border cells involved in the secretion of molecules required for efficient root interaction with the environment, and ala3 mutants are devoid of the characteristic trans-Golgi proliferation of slime vesicles containing polysaccharides and enzymes for secretion. In yeast complementation experiments, ALA3 function requires interaction with members of a novel family of plant membrane-bound proteins, ALIS1 to ALIS5 (for ALA-Interacting Subunit), and in this host ALA3 and ALIS1 show strong affinity for each other. In planta, ALIS1, like ALA3, localizes to Golgi-like structures and is expressed in root peripheral columella cells. We propose that the ALIS1 protein is a beta-subunit of ALA3 and that this protein complex forms an important part of the Golgi machinery required for secretory processes during plant development.
Figures





Similar articles
-
Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate.Plant J. 2012 Aug;71(3):479-91. doi: 10.1111/j.1365-313X.2012.05004.x. Epub 2012 May 25. Plant J. 2012. PMID: 22449068
-
The Tip-Localized Phosphatidylserine Established by Arabidopsis ALA3 Is Crucial for Rab GTPase-Mediated Vesicle Trafficking and Pollen Tube Growth.Plant Cell. 2020 Oct;32(10):3170-3187. doi: 10.1105/tpc.19.00844. Epub 2020 Aug 18. Plant Cell. 2020. PMID: 32817253 Free PMC article.
-
Loss of the Arabidopsis thaliana P₄-ATPase ALA3 reduces adaptability to temperature stresses and impairs vegetative, pollen, and ovule development.PLoS One. 2013 May 7;8(5):e62577. doi: 10.1371/journal.pone.0062577. Print 2013. PLoS One. 2013. PMID: 23667493 Free PMC article.
-
Links between lipid homeostasis, organelle morphodynamics and protein trafficking in eukaryotic and plant secretory pathways.Plant Cell Rep. 2011 Feb;30(2):177-93. doi: 10.1007/s00299-010-0954-1. Epub 2010 Dec 1. Plant Cell Rep. 2011. PMID: 21120657 Review.
-
The Plant V-ATPase.Front Plant Sci. 2022 Jun 30;13:931777. doi: 10.3389/fpls.2022.931777. eCollection 2022. Front Plant Sci. 2022. PMID: 35845650 Free PMC article. Review.
Cited by
-
A potential pathway for flippase-facilitated glucosylceramide catabolism in plants.Plant Signal Behav. 2020 Oct 2;15(10):1783486. doi: 10.1080/15592324.2020.1783486. Epub 2020 Aug 28. Plant Signal Behav. 2020. PMID: 32857675 Free PMC article.
-
The lipid head group is the key element for substrate recognition by the P4 ATPase ALA2: a phosphatidylserine flippase.Biochem J. 2019 Mar 6;476(5):783-794. doi: 10.1042/BCJ20180891. Biochem J. 2019. PMID: 30755463 Free PMC article.
-
Mechanisms of membrane curvature generation in membrane traffic.Membranes (Basel). 2012 Feb 29;2(1):118-33. doi: 10.3390/membranes2010118. Membranes (Basel). 2012. PMID: 24957965 Free PMC article.
-
Lipid somersaults: Uncovering the mechanisms of protein-mediated lipid flipping.Prog Lipid Res. 2016 Oct;64:69-84. doi: 10.1016/j.plipres.2016.08.003. Epub 2016 Aug 12. Prog Lipid Res. 2016. PMID: 27528189 Free PMC article. Review.
-
Direct evidence of lipid transport by the Drs2-Cdc50 flippase upon truncation of its terminal regions.Protein Sci. 2023 Dec 8;33(3):e4855. doi: 10.1002/pro.4855. Online ahead of print. Protein Sci. 2023. PMID: 38063271 Free PMC article.
References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Altendorf, K., Gassel, M., Puppe, W., Mollenkamp, T., Zeeck, A., Boddien, C., Fendler, K., Bamberg, E., and Drose, S. (1998). Structure and function of the Kdp-ATPase of Escherichia coli. Acta Physiol. Scand. Suppl. 643 137–146. - PubMed
-
- Bolwell, G.P. (1988). Synthesis of cell wall components: Aspects of control. Phytochemistry 27 1235–1253.
-
- Brighman, L.A., Woo, H.H., and Hawes, M.C. (1995). Root border cells as tools in plant cell studies. Methods Cell Biol. 49 377–387. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

