Following translation by single ribosomes one codon at a time
- PMID: 18327250
- PMCID: PMC2556548
- DOI: 10.1038/nature06716
Following translation by single ribosomes one codon at a time
Abstract
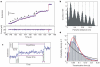
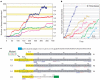
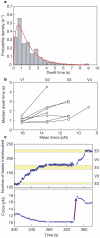
We have followed individual ribosomes as they translate single messenger RNA hairpins tethered by the ends to optical tweezers. Here we reveal that translation occurs through successive translocation--and-pause cycles. The distribution of pause lengths, with a median of 2.8 s, indicates that at least two rate-determining processes control each pause. Each translocation step measures three bases--one codon-and occurs in less than 0.1 s. Analysis of the times required for translocation reveals, surprisingly, that there are three substeps in each step. Pause lengths, and thus the overall rate of translation, depend on the secondary structure of the mRNA; the applied force destabilizes secondary structure and decreases pause durations, but does not affect translocation times. Translocation and RNA unwinding are strictly coupled ribosomal functions.
Figures

Similar articles
-
Real-time tRNA transit on single translating ribosomes at codon resolution.Nature. 2010 Apr 15;464(7291):1012-7. doi: 10.1038/nature08925. Nature. 2010. PMID: 20393556 Free PMC article.
-
The ribosome uses two active mechanisms to unwind messenger RNA during translation.Nature. 2011 Jul 6;475(7354):118-21. doi: 10.1038/nature10126. Nature. 2011. PMID: 21734708 Free PMC article.
-
Direct measurement of the mechanical work during translocation by the ribosome.Elife. 2014 Aug 11;3:e03406. doi: 10.7554/eLife.03406. Elife. 2014. PMID: 25114092 Free PMC article.
-
Meanderings of the mRNA through the ribosome.Structure. 2001 Sep;9(9):751-8. doi: 10.1016/s0969-2126(01)00649-9. Structure. 2001. PMID: 11566123 Review.
-
Probing the mechanisms of translation with force.Chem Rev. 2014 Mar 26;114(6):3266-80. doi: 10.1021/cr400313x. Epub 2014 Jan 9. Chem Rev. 2014. PMID: 24405158 Free PMC article. Review. No abstract available.
Cited by
-
Torque measurement at the single-molecule level.Annu Rev Biophys. 2013;42:583-604. doi: 10.1146/annurev-biophys-083012-130412. Annu Rev Biophys. 2013. PMID: 23541162 Free PMC article. Review.
-
Dynamics of translation by single ribosomes through mRNA secondary structures.Nat Struct Mol Biol. 2013 May;20(5):582-8. doi: 10.1038/nsmb.2544. Epub 2013 Mar 31. Nat Struct Mol Biol. 2013. PMID: 23542154 Free PMC article.
-
Resistance of mRNAs with AUG-proximal nonsense mutations to nonsense-mediated decay reflects variables of mRNA structure and translational activity.Nucleic Acids Res. 2015 Jul 27;43(13):6528-44. doi: 10.1093/nar/gkv588. Epub 2015 Jun 11. Nucleic Acids Res. 2015. PMID: 26068473 Free PMC article.
-
Nonfluorescent quenchers to correlate single-molecule conformational and compositional dynamics.J Am Chem Soc. 2012 Apr 4;134(13):5734-7. doi: 10.1021/ja2119964. Epub 2012 Mar 23. J Am Chem Soc. 2012. PMID: 22428667 Free PMC article.
-
The ribosome in action: Tuning of translational efficiency and protein folding.Protein Sci. 2016 Aug;25(8):1390-406. doi: 10.1002/pro.2950. Epub 2016 Jun 8. Protein Sci. 2016. PMID: 27198711 Free PMC article. Review.
References
-
- Smith SB, Cui Y, Bustamante C. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 2003;361:134–162. - PubMed
-
- Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. - PubMed
-
- Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005;20:623–632. - PubMed
-
- Wilson KS, Noller HF. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell. 1998;92:131–139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

