Moonlighting proteins in yeasts
- PMID: 18322039
- PMCID: PMC2268286
- DOI: 10.1128/MMBR.00036-07
Moonlighting proteins in yeasts
Abstract
Proteins able to participate in unrelated biological processes have been grouped under the generic name of moonlighting proteins. Work with different yeast species has uncovered a great number of moonlighting proteins and shown their importance for adequate functioning of the yeast cell. Moonlighting activities in yeasts include such diverse functions as control of gene expression, organelle assembly, and modification of the activity of metabolic pathways. In this review, we consider several well-studied moonlighting proteins in different yeast species, paying attention to the experimental approaches used to identify them and the evidence that supports their participation in the unexpected function. Usually, moonlighting activities have been uncovered unexpectedly, and up to now, no satisfactory way to predict moonlighting activities has been found. Among the well-characterized moonlighting proteins in yeasts, enzymes from the glycolytic pathway appear to be prominent. For some cases, it is shown that despite close phylogenetic relationships, moonlighting activities are not necessarily conserved among yeast species. Organisms may utilize moonlighting to add a new layer of regulation to conventional regulatory networks. The existence of this type of proteins in yeasts should be taken into account when designing mutant screens or in attempts to model or modify yeast metabolism.
Figures
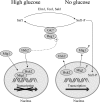
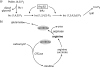
Similar articles
-
Unraveling moonlighting functions with yeasts.IUBMB Life. 2011 Jul;63(7):457-62. doi: 10.1002/iub.454. Epub 2011 Apr 13. IUBMB Life. 2011. PMID: 21491559 Review.
-
The Expanding Landscape of Moonlighting Proteins in Yeasts.Microbiol Mol Biol Rev. 2016 Jul 27;80(3):765-77. doi: 10.1128/MMBR.00012-16. Print 2016 Sep. Microbiol Mol Biol Rev. 2016. PMID: 27466281 Free PMC article. Review.
-
Structural analysis of N-/O-glycans assembled on proteins in yeasts.J Microbiol. 2018 Jan;56(1):11-23. doi: 10.1007/s12275-018-7468-x. Epub 2018 Jan 4. J Microbiol. 2018. PMID: 29299842
-
[Iron metabolism in the yeast].Ukr Biokhim Zh (1999). 2005 May-Jun;77(3):5-19. Ukr Biokhim Zh (1999). 2005. PMID: 16566123 Review. Ukrainian.
-
Yeast expression platforms.Appl Microbiol Biotechnol. 2007 Dec;77(3):513-23. doi: 10.1007/s00253-007-1209-0. Epub 2007 Oct 9. Appl Microbiol Biotechnol. 2007. PMID: 17924105 Review.
Cited by
-
Protein dynamics and the diversity of an antibody response.J Biol Chem. 2012 Aug 3;287(32):27139-47. doi: 10.1074/jbc.M112.372698. Epub 2012 Jun 8. J Biol Chem. 2012. PMID: 22685303 Free PMC article.
-
Evaluation of function predictions by PFP, ESG,and PSI-BLAST for moonlighting proteins.BMC Proc. 2012 Nov 13;6 Suppl 7(Suppl 7):S5. doi: 10.1186/1753-6561-6-S7-S5. Epub 2012 Nov 13. BMC Proc. 2012. PMID: 23173871 Free PMC article.
-
Protein promiscuity and its implications for biotechnology.Nat Biotechnol. 2009 Feb;27(2):157-67. doi: 10.1038/nbt1519. Nat Biotechnol. 2009. PMID: 19204698 Review.
-
Surface-associated plasminogen binding of Cryptococcus neoformans promotes extracellular matrix invasion.PLoS One. 2009 Jun 3;4(6):e5780. doi: 10.1371/journal.pone.0005780. PLoS One. 2009. PMID: 19492051 Free PMC article.
-
Physical Features of Intracellular Proteins that Moonlight on the Cell Surface.PLoS One. 2015 Jun 25;10(6):e0130575. doi: 10.1371/journal.pone.0130575. eCollection 2015. PLoS One. 2015. PMID: 26110848 Free PMC article.
References
-
- Ahuatzi, D., P. Herrero, T. de la Cera, and F. Moreno. 2004. The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 27914440-14446. - PubMed
-
- Ahuatzi, D., A. Riera, R. Pelaez, P. Herrero, and F. Moreno. 2007. Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J. Biol. Chem. 2824485-4493. - PubMed
-
- Aigle, M., and F. Lacroute. 1975. Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol. Gen. Genet. 136327-335. - PubMed
-
- Anders, A., H. Lilie, K. Franke, L. Kapp, J. Stelling, E. D. Gilles, and K. D. Breunig. 2006. The galactose switch in Kluyveromyces lactis depends on nuclear competition between Gal4 and Gal1 for Gal80 binding. J. Biol. Chem. 28129337-29348. - PubMed
-
- Armstrong, R. N. 1997. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 102-18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

