NF-kappaB-mediated activation of the chemokine CCL22 by the product of the human cytomegalovirus gene UL144 escapes regulation by viral IE86
- PMID: 18287226
- PMCID: PMC2293074
- DOI: 10.1128/JVI.02156-07
NF-kappaB-mediated activation of the chemokine CCL22 by the product of the human cytomegalovirus gene UL144 escapes regulation by viral IE86
Abstract
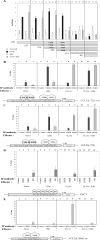
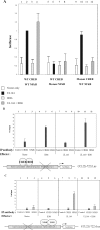
The product of the human cytomegalovirus (HCMV) gene UL144, expressed at early times postinfection, is located in the UL/b' region of the viral genome and is related to members of the tumor necrosis factor receptor superfamily, but it does not bind tumor necrosis factor superfamily ligands. However, UL144 does activate NF-kappaB, resulting in NF-kappaB-mediated activation of the cellular chemokine CCL22. Consistent with this finding, isolates of HCMV lacking the UL/b' region show no such activation of CCL22. Recently, it has been suggested that activation of NF-kappaB is repressed by the product of the viral gene IE86: IE86 appears to block NF-kappaB binding to DNA but not nuclear translocation of NF-kappaB. Intriguingly, IE86 is detectable throughout an infection with the virus, so how UL144 is able to activate NF-kappaB in the presence of continued IE86 expression is unclear. Here we show that although IE86 does repress the UL144-mediated activation of a synthetic NF-kappaB promoter, it is unable to block UL144-mediated activation of the CCL22 promoter, and this lack of responsiveness to IE86 appears to be regulated by binding of the CREB transcription factor.
Figures

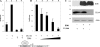

Similar articles
-
Identification of TRIM23 as a cofactor involved in the regulation of NF-kappaB by human cytomegalovirus.J Virol. 2009 Apr;83(8):3581-90. doi: 10.1128/JVI.02072-08. Epub 2009 Jan 28. J Virol. 2009. PMID: 19176615 Free PMC article.
-
The UL144 gene product of human cytomegalovirus activates NFkappaB via a TRAF6-dependent mechanism.EMBO J. 2006 Sep 20;25(18):4390-9. doi: 10.1038/sj.emboj.7601287. Epub 2006 Aug 24. EMBO J. 2006. PMID: 16932746 Free PMC article.
-
Human cytomegalovirus IE86 attenuates virus- and tumor necrosis factor alpha-induced NFkappaB-dependent gene expression.J Virol. 2006 Nov;80(21):10763-71. doi: 10.1128/JVI.01195-06. J Virol. 2006. PMID: 17041226 Free PMC article.
-
Functional roles of the human cytomegalovirus essential IE86 protein.Curr Top Microbiol Immunol. 2008;325:133-52. doi: 10.1007/978-3-540-77349-8_8. Curr Top Microbiol Immunol. 2008. PMID: 18637504 Review.
-
The power of human cytomegalovirus (HCMV) hijacked UL/b' functions lost in vitro.Acta Virol. 2020;64(2):117-130. doi: 10.4149/av_2020_202. Acta Virol. 2020. PMID: 32551781 Review.
Cited by
-
The myeloid transcription factor GATA-2 regulates the viral UL144 gene during human cytomegalovirus latency in an isolate-specific manner.J Virol. 2013 Apr;87(8):4261-71. doi: 10.1128/JVI.03497-12. Epub 2013 Jan 30. J Virol. 2013. PMID: 23365437 Free PMC article.
-
Identification of TRIM23 as a cofactor involved in the regulation of NF-kappaB by human cytomegalovirus.J Virol. 2009 Apr;83(8):3581-90. doi: 10.1128/JVI.02072-08. Epub 2009 Jan 28. J Virol. 2009. PMID: 19176615 Free PMC article.
-
Aflatoxin B1 and Epstein-Barr virus-induced CCL22 expression stimulates B cell infection.Proc Natl Acad Sci U S A. 2024 Apr 16;121(16):e2314426121. doi: 10.1073/pnas.2314426121. Epub 2024 Apr 4. Proc Natl Acad Sci U S A. 2024. PMID: 38574017 Free PMC article.
-
Human cytomegalovirus encoded homologs of cytokines, chemokines and their receptors: roles in immunomodulation.Viruses. 2012 Oct 25;4(11):2448-70. doi: 10.3390/v4112448. Viruses. 2012. PMID: 23202490 Free PMC article. Review.
-
Who's Driving? Human Cytomegalovirus, Interferon, and NFκB Signaling.Viruses. 2018 Aug 21;10(9):447. doi: 10.3390/v10090447. Viruses. 2018. PMID: 30134546 Free PMC article. Review.
References
-
- Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 27439-55. - PubMed
-
- Baracchini, E., E. Glezer, K. Fish, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1992. An isoform variant of the cytomegalovirus immediate-early auto repressor functions as a transcriptional activator. Virology 188518-519. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

