DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells
- PMID: 18285465
- PMCID: PMC2293113
- DOI: 10.1128/MCB.02076-07
DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells
Abstract
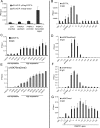
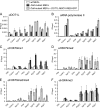
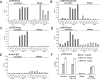
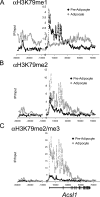
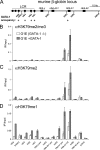
The histone H3 lysine 79 methyltransferase DOT1L/KMT4 can promote an oncogenic pattern of gene expression through binding with several MLL fusion partners found in acute leukemia. However, the normal function of DOT1L in mammalian gene regulation is poorly understood. Here we report that DOT1L recruitment is ubiquitously coupled with active transcription in diverse mammalian cell types. DOT1L preferentially occupies the proximal transcribed region of active genes, correlating with enrichment of H3K79 di- and trimethylation. Furthermore, Dot1l mutant fibroblasts lacked H3K79 di- and trimethylation at all sites examined, indicating that DOT1L is the sole enzyme responsible for these marks. Importantly, we identified chromatin immunoprecipitation (ChIP) assay conditions necessary for reliable H3K79 methylation detection. ChIP-chip tiling arrays revealed that levels of all degrees of genic H3K79 methylation correlate with mRNA abundance and dynamically respond to changes in gene activity. Conversion of H3K79 monomethylation into di- and trimethylation correlated with the transition from low- to high-level gene transcription. We also observed enrichment of H3K79 monomethylation at intergenic regions occupied by DNA-binding transcriptional activators. Our findings highlight several similarities between the patterning of H3K4 methylation and that of H3K79 methylation in mammalian chromatin, suggesting a widespread mechanism for parallel or sequential recruitment of DOT1L and MLL to genes in their normal "on" state.
Figures



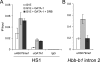
Similar articles
-
The lncRNA LAMP5-AS1 drives leukemia cell stemness by directly modulating DOT1L methyltransferase activity in MLL leukemia.J Hematol Oncol. 2020 Jun 17;13(1):78. doi: 10.1186/s13045-020-00909-y. J Hematol Oncol. 2020. PMID: 32552847 Free PMC article.
-
The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure.PLoS Genet. 2008 Sep 12;4(9):e1000190. doi: 10.1371/journal.pgen.1000190. PLoS Genet. 2008. PMID: 18787701 Free PMC article.
-
The upstreams and downstreams of H3K79 methylation by DOT1L.Chromosoma. 2016 Sep;125(4):593-605. doi: 10.1007/s00412-015-0570-5. Epub 2016 Jan 4. Chromosoma. 2016. PMID: 26728620 Review.
-
DOT1L-mediated H3K79 methylation in chromatin is dispensable for Wnt pathway-specific and other intestinal epithelial functions.Mol Cell Biol. 2013 May;33(9):1735-45. doi: 10.1128/MCB.01463-12. Epub 2013 Feb 19. Mol Cell Biol. 2013. PMID: 23428873 Free PMC article.
-
The histone methyltransferase Dot1/DOT1L as a critical regulator of the cell cycle.Cell Cycle. 2014;13(5):726-38. doi: 10.4161/cc.28104. Epub 2014 Feb 6. Cell Cycle. 2014. PMID: 24526115 Free PMC article. Review.
Cited by
-
DOT1L promotes angiogenesis through cooperative regulation of VEGFR2 with ETS-1.Oncotarget. 2016 Oct 25;7(43):69674-69687. doi: 10.18632/oncotarget.11939. Oncotarget. 2016. PMID: 27626484 Free PMC article.
-
A Conserved Nuclear Cyclophilin Is Required for Both RNA Polymerase II Elongation and Co-transcriptional Splicing in Caenorhabditis elegans.PLoS Genet. 2016 Aug 19;12(8):e1006227. doi: 10.1371/journal.pgen.1006227. eCollection 2016 Aug. PLoS Genet. 2016. PMID: 27541139 Free PMC article.
-
The lncRNA LAMP5-AS1 drives leukemia cell stemness by directly modulating DOT1L methyltransferase activity in MLL leukemia.J Hematol Oncol. 2020 Jun 17;13(1):78. doi: 10.1186/s13045-020-00909-y. J Hematol Oncol. 2020. PMID: 32552847 Free PMC article.
-
Targeting epigenetic regulators for inflammation: Mechanisms and intervention therapy.MedComm (2020). 2022 Sep 15;3(4):e173. doi: 10.1002/mco2.173. eCollection 2022 Dec. MedComm (2020). 2022. PMID: 36176733 Free PMC article. Review.
-
IKAROS is required for the measured response of NOTCH target genes upon external NOTCH signaling.PLoS Genet. 2021 Mar 26;17(3):e1009478. doi: 10.1371/journal.pgen.1009478. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33770102 Free PMC article.
References
-
- Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. - PubMed
-
- Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120169-181. - PubMed
-
- Bernstein, B. E., T. S. Mikkelsen, X. Xie, M. Kamal, D. J. Huebert, J. Cuff, B. Fry, A. Meissner, M. Wernig, K. Plath, R. Jaenisch, A. Wagschal, R. Feil, S. L. Schreiber, and E. S. Lander. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125315-326. - PubMed
-
- Bitoun, E., P. L. Oliver, and K. E. Davies. 2007. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 1692-106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
