Messenger RNA regulation: to translate or to degrade
- PMID: 18256698
- PMCID: PMC2241649
- DOI: 10.1038/sj.emboj.7601977
Messenger RNA regulation: to translate or to degrade
Abstract
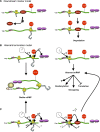
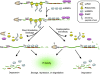
Quality control of gene expression operates post-transcriptionally at various levels in eukaryotes. Once transcribed, mRNAs associate with a host of proteins throughout their lifetime. These mRNA-protein complexes (mRNPs) undergo a series of remodeling events that are influenced by and/or influence the translation and mRNA decay machinery. In this review we discuss how a decision to translate or to degrade a cytoplasmic mRNA is reached. Nonsense-mediated mRNA decay (NMD) and microRNA (miRNA)-mediated mRNA silencing are provided as examples. NMD is a surveillance mechanism that detects and eliminates aberrant mRNAs whose expression would result in truncated proteins that are often deleterious to the organism. miRNA-mediated mRNA silencing is a mechanism that ensures a given protein is expressed at a proper level to permit normal cellular function. While NMD and miRNA-mediated mRNA silencing use different decision-making processes to determine the fate of their targets, both are greatly influenced by mRNP dynamics. In addition, both are linked to RNA processing bodies. Possible modes involving 3' untranslated region and its associated factors, which appear to play key roles in both processes, are discussed.
Figures
Similar articles
-
[Nonsense-mediated mRNA decay (NMD)--on guard of mRNA quality].Postepy Biochem. 2006;52(4):390-8. Postepy Biochem. 2006. PMID: 17536508 Review. Polish.
-
Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast.PLoS Genet. 2006 Nov 24;2(11):e203. doi: 10.1371/journal.pgen.0020203. Epub 2006 Oct 18. PLoS Genet. 2006. PMID: 17166056 Free PMC article.
-
NMD: multitasking between mRNA surveillance and modulation of gene expression.Adv Genet. 2008;62:185-243. doi: 10.1016/S0065-2660(08)00604-4. Adv Genet. 2008. PMID: 19010255 Review.
-
P-bodies and stress granules: possible roles in the control of translation and mRNA degradation.Cold Spring Harb Perspect Biol. 2012 Sep 1;4(9):a012286. doi: 10.1101/cshperspect.a012286. Cold Spring Harb Perspect Biol. 2012. PMID: 22763747 Free PMC article. Review.
-
microRNA/Argonaute 2 regulates nonsense-mediated messenger RNA decay.EMBO Rep. 2010 May;11(5):380-6. doi: 10.1038/embor.2010.44. Epub 2010 Apr 16. EMBO Rep. 2010. PMID: 20395958 Free PMC article.
Cited by
-
Antisense technology: an emerging platform for cardiovascular disease therapeutics.J Cardiovasc Transl Res. 2013 Dec;6(6):969-80. doi: 10.1007/s12265-013-9495-7. Epub 2013 Jul 16. J Cardiovasc Transl Res. 2013. PMID: 23856914 Free PMC article. Review.
-
Protein segregase meddles in remodeling of mRNA-protein complexes.Genes Dev. 2013 May 1;27(9):980-4. doi: 10.1101/gad.219469.113. Genes Dev. 2013. PMID: 23651853 Free PMC article.
-
Mobility of Transgenic Nucleic Acids and Proteins within Grafted Rootstocks for Agricultural Improvement.Front Plant Sci. 2012 Mar 2;3:39. doi: 10.3389/fpls.2012.00039. eCollection 2012. Front Plant Sci. 2012. PMID: 22645583 Free PMC article.
-
When a ribosome encounters a premature termination codon.BMB Rep. 2013 Jan;46(1):9-16. doi: 10.5483/bmbrep.2013.46.1.002. BMB Rep. 2013. PMID: 23351378 Free PMC article. Review.
-
The role of microRNAs miR-200b and miR-200c in TLR4 signaling and NF-κB activation.Innate Immun. 2012 Dec;18(6):846-55. doi: 10.1177/1753425912443903. Epub 2012 Apr 20. Innate Immun. 2012. PMID: 22522429 Free PMC article.
References
-
- Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355 - PubMed
-
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 - PubMed
-
- Amrani N, Sachs MS, Jacobson A (2006) Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 7: 415–425 - PubMed
-
- Arciga-Reyes L, Wootton L, Kieffer M, Davies B (2006) UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J 47: 480–489 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

