Structure-function analysis of vaccinia virus mRNA cap (guanine-N7) methyltransferase
- PMID: 18256245
- PMCID: PMC2271365
- DOI: 10.1261/rna.928208
Structure-function analysis of vaccinia virus mRNA cap (guanine-N7) methyltransferase
Abstract
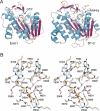
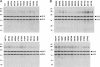
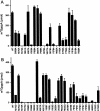
The guanine-N7 methyltransferase domain of vaccinia virus mRNA capping enzyme is a heterodimer composed of a catalytic subunit and a stimulatory subunit. Structure-function analysis of the catalytic subunit by alanine scanning and conservative substitutions (49 mutations at 25 amino acids) identified 12 functional groups essential for methyltransferase activity in vivo, most of which were essential for cap methylation in vitro. Defects in cap binding were demonstrated for a subset of lethal mutants that displayed residual activity in vitro. We discuss our findings in light of a model of the Michaelis complex derived from crystal structures of AdoHcy-bound vaccinia cap methyltransferase and GTP-bound cellular cap methyltransferase. The structure-function data yield a coherent picture of the vaccinia cap methyltransferase active site and the determinants of substrate specificity and affinity.
Figures
Similar articles
-
Mutational analysis of vaccinia virus mRNA cap (guanine-N7) methyltransferase reveals essential contributions of the N-terminal peptide that closes over the active site.RNA. 2008 Nov;14(11):2297-304. doi: 10.1261/rna.1201308. Epub 2008 Sep 17. RNA. 2008. PMID: 18799596 Free PMC article.
-
Vaccinia virus mRNA (guanine-7-)methyltransferase: mutational effects on cap methylation and AdoHcy-dependent photo-cross-linking of the cap to the methyl acceptor site.Biochemistry. 1996 May 28;35(21):6900-10. doi: 10.1021/bi960221a. Biochemistry. 1996. PMID: 8639642
-
Effects of alanine cluster mutations in the D12 subunit of vaccinia virus mRNA (guanine-N7) methyltransferase.Virology. 2001 Aug 15;287(1):40-8. doi: 10.1006/viro.2001.1006. Virology. 2001. PMID: 11504540
-
Flavivirus methyltransferase: a novel antiviral target.Antiviral Res. 2008 Oct;80(1):1-10. doi: 10.1016/j.antiviral.2008.05.003. Epub 2008 Jun 5. Antiviral Res. 2008. PMID: 18571739 Free PMC article. Review.
-
Regulation of mRNA capping in the cell cycle.RNA Biol. 2017 Jan 2;14(1):11-14. doi: 10.1080/15476286.2016.1251540. Epub 2016 Oct 28. RNA Biol. 2017. PMID: 27791484 Free PMC article. Review.
Cited by
-
Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs.Proc Natl Acad Sci U S A. 2010 Aug 17;107(33):14787-92. doi: 10.1073/pnas.1009490107. Epub 2010 Aug 2. Proc Natl Acad Sci U S A. 2010. PMID: 20679221 Free PMC article.
-
Versatile strategy using vaccinia virus-capping enzyme to synthesize functional 5' cap-modified mRNAs.Nucleic Acids Res. 2023 Apr 11;51(6):e34. doi: 10.1093/nar/gkad019. Nucleic Acids Res. 2023. PMID: 36731515 Free PMC article.
-
Crystal structure of vaccinia virus mRNA capping enzyme provides insights into the mechanism and evolution of the capping apparatus.Structure. 2014 Mar 4;22(3):452-65. doi: 10.1016/j.str.2013.12.014. Structure. 2014. PMID: 24607143 Free PMC article.
-
Structure-function analysis of severe acute respiratory syndrome coronavirus RNA cap guanine-N7-methyltransferase.J Virol. 2013 Jun;87(11):6296-305. doi: 10.1128/JVI.00061-13. Epub 2013 Mar 27. J Virol. 2013. PMID: 23536667 Free PMC article.
-
Mutational analysis of vaccinia virus mRNA cap (guanine-N7) methyltransferase reveals essential contributions of the N-terminal peptide that closes over the active site.RNA. 2008 Nov;14(11):2297-304. doi: 10.1261/rna.1201308. Epub 2008 Sep 17. RNA. 2008. PMID: 18799596 Free PMC article.
References
-
- Chrebet, G.L., Wisniewski, D., Perkins, A.L., Deng, Q., Kurtz, M.B., Marcy, A., Parent, S.A. Cell-based assays to detect inhibitors of fungal mRNA capping enzymes and characterization of sinefungin as a cap methyltransferase inhibitor. J. Biomol. Screen. 2005;10:355–364. - PubMed
-
- Cong, P., Shuman, S. Methyltransferase and subunit association domains of vaccinia virus mRNA capping enzyme. J. Biol. Chem. 1992;267:16424–16429. - PubMed
-
- Fabrega, C., Hausmann, S., Shen, V., Shuman, S., Lima, C.D. Structure and mechanism of cap (guanine-N7) methyltransferase. Mol. Cell. 2004;13:77–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
