The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels
- PMID: 18252845
- PMCID: PMC2276449
- DOI: 10.1105/tpc.107.052142
The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels
Abstract
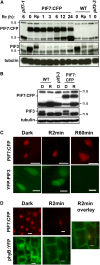
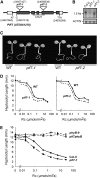
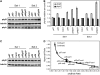
We show that a previously uncharacterized Arabidopsis thaliana basic helix-loop-helix (bHLH) phytochrome interacting factor (PIF), designated PIF7, interacts specifically with the far-red light-absorbing Pfr form of phyB through a conserved domain called the active phyB binding motif. Similar to PIF3, upon light exposure, PIF7 rapidly migrates to intranuclear speckles, where it colocalizes with phyB. However, in striking contrast to PIF3, this process is not accompanied by detectable light-induced phosphorylation or degradation of PIF7, suggesting that the consequences of interaction with photoactivated phyB may differ among PIFs. Nevertheless, PIF7 acts similarly to PIF3 in prolonged red light as a weak negative regulator of phyB-mediated seedling deetiolation. Examination of pif3, pif4, and pif7 double mutant combinations shows that their moderate hypersensitivity to extended red light is additive. We provide evidence that the mechanism by which these PIFs operate on the phyB signaling pathway under prolonged red light is through maintaining low phyB protein levels, in an additive or synergistic manner, via a process likely involving the proteasome pathway. These data suggest that the role of these phyB-interacting bHLH factors in modulating seedling deetiolation in prolonged red light may not be as phy-activated signaling intermediates, as proposed previously, but as direct modulators of the abundance of the photoreceptor.
Figures

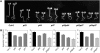

Similar articles
-
Phytochrome signaling in green Arabidopsis seedlings: impact assessment of a mutually negative phyB-PIF feedback loop.Mol Plant. 2012 May;5(3):734-49. doi: 10.1093/mp/sss031. Epub 2012 Apr 5. Mol Plant. 2012. PMID: 22492120 Free PMC article.
-
Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes.Plant Cell. 2008 Jun;20(6):1586-602. doi: 10.1105/tpc.108.060020. Epub 2008 Jun 6. Plant Cell. 2008. PMID: 18539749 Free PMC article.
-
Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis.Plant Cell. 2013 Jul;25(7):2679-98. doi: 10.1105/tpc.113.112342. Epub 2013 Jul 31. Plant Cell. 2013. PMID: 23903316 Free PMC article.
-
Phytochrome-interacting factor from Arabidopsis to liverwort.Curr Opin Plant Biol. 2017 Feb;35:54-60. doi: 10.1016/j.pbi.2016.11.004. Epub 2016 Nov 19. Curr Opin Plant Biol. 2017. PMID: 27875778 Review.
-
PIFs: systems integrators in plant development.Plant Cell. 2014 Jan;26(1):56-78. doi: 10.1105/tpc.113.120857. Epub 2014 Jan 30. Plant Cell. 2014. PMID: 24481072 Free PMC article. Review.
Cited by
-
Shade avoidance.Arabidopsis Book. 2012;10:e0157. doi: 10.1199/tab.0157. Epub 2012 Jan 19. Arabidopsis Book. 2012. PMID: 22582029 Free PMC article.
-
SPAs promote thermomorphogenesis by regulating the phyB-PIF4 module in Arabidopsis.Development. 2020 Oct 8;147(19):dev189233. doi: 10.1242/dev.189233. Development. 2020. PMID: 32994167 Free PMC article.
-
Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis.Genes Dev. 2012 Aug 15;26(16):1851-63. doi: 10.1101/gad.193219.112. Genes Dev. 2012. PMID: 22895253 Free PMC article.
-
Regulation of Carotenoid Biosynthesis by Shade Relies on Specific Subsets of Antagonistic Transcription Factors and Cofactors.Plant Physiol. 2015 Nov;169(3):1584-94. doi: 10.1104/pp.15.00552. Epub 2015 Jun 16. Plant Physiol. 2015. PMID: 26082398 Free PMC article.
-
Phylogeny and evolution of plant Phytochrome Interacting Factors (PIFs) gene family and functional analyses of PIFs in Brachypodium distachyon.Plant Cell Rep. 2022 May;41(5):1209-1227. doi: 10.1007/s00299-022-02850-5. Epub 2022 Feb 26. Plant Cell Rep. 2022. PMID: 35218399
References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Al-Sady, B., Ni, W., Kircher, S., Schafer, E., and Quail, P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation as a prelude to proteasome-mediated degradation. Mol. Cell 23 439–446. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

