SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway
- PMID: 18227861
- PMCID: PMC7091891
- DOI: 10.1038/cr.2008.15
SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway
Abstract
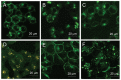
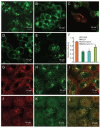
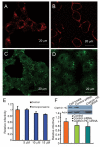
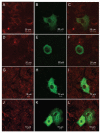
While severe acute respiratory syndrome coronavirus (SARS-CoV) was initially thought to enter cells through direct fusion with the plasma membrane, more recent evidence suggests that virus entry may also involve endocytosis. We have found that SARS-CoV enters cells via pH- and receptor-dependent endocytosis. Treatment of cells with either SARS-CoV spike protein or spike-bearing pseudoviruses resulted in the translocation of angiotensin-converting enzyme 2 (ACE2), the functional receptor of SARS-CoV, from the cell surface to endosomes. In addition, the spike-bearing pseudoviruses and early endosome antigen 1 were found to colocalize in endosomes. Further analyses using specific endocytic pathway inhibitors and dominant-negative Eps15 as well as caveolin-1 colocalization study suggested that virus entry was mediated by a clathrin- and caveolae-independent mechanism. Moreover, cholesterol- and sphingolipid-rich lipid raft microdomains in the plasma membrane, which have been shown to act as platforms for many physiological signaling pathways, were shown to be involved in virus entry. Endocytic entry of SARS-CoV may expand the cellular range of SARS-CoV infection, and our findings here contribute to the understanding of SARS-CoV pathogenesis, providing new information for anti-viral drug research.
Figures
Similar articles
-
Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted.J Virol. 2007 Aug;81(16):8722-9. doi: 10.1128/JVI.00253-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522231 Free PMC article.
-
Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2.Virology. 2008 Nov 25;381(2):215-21. doi: 10.1016/j.virol.2008.08.026. Epub 2008 Sep 23. Virology. 2008. PMID: 18814896 Free PMC article.
-
Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry.Proc Natl Acad Sci U S A. 2008 Jun 3;105(22):7809-14. doi: 10.1073/pnas.0711241105. Epub 2008 May 19. Proc Natl Acad Sci U S A. 2008. PMID: 18490652 Free PMC article.
-
Severe acute respiratory syndrome coronavirus entry as a target of antiviral therapies.Antivir Ther. 2007;12(4 Pt B):639-50. Antivir Ther. 2007. PMID: 17944271 Review.
-
Cellular entry of the SARS coronavirus.Trends Microbiol. 2004 Oct;12(10):466-72. doi: 10.1016/j.tim.2004.08.008. Trends Microbiol. 2004. PMID: 15381196 Free PMC article. Review.
Cited by
-
Various Facets of Pathogenic Lipids in Infectious Diseases: Exploring Virulent Lipid-Host Interactome and Their Druggability.J Membr Biol. 2020 Oct;253(5):399-423. doi: 10.1007/s00232-020-00135-0. Epub 2020 Aug 24. J Membr Biol. 2020. PMID: 32833058 Free PMC article. Review.
-
Sex differences underlying preexisting cardiovascular disease and cardiovascular injury in COVID-19.J Mol Cell Cardiol. 2020 Nov;148:25-33. doi: 10.1016/j.yjmcc.2020.08.007. Epub 2020 Aug 22. J Mol Cell Cardiol. 2020. PMID: 32835666 Free PMC article.
-
Can cilia provide an entry gateway for SARS-CoV-2 to human ciliated cells?Physiol Genomics. 2021 Jun 1;53(6):249-258. doi: 10.1152/physiolgenomics.00015.2021. Epub 2021 Apr 15. Physiol Genomics. 2021. PMID: 33855870 Free PMC article. Review.
-
The recent outbreaks of human coronaviruses: A medicinal chemistry perspective.Med Res Rev. 2021 Jan;41(1):72-135. doi: 10.1002/med.21724. Epub 2020 Aug 27. Med Res Rev. 2021. PMID: 32852058 Free PMC article. Review.
-
Insight into the role of clathrin-mediated endocytosis inhibitors in SARS-CoV-2 infection.Rev Med Virol. 2023 Jan;33(1):e2403. doi: 10.1002/rmv.2403. Epub 2022 Nov 7. Rev Med Virol. 2023. PMID: 36345157 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

