Chp1-Tas3 interaction is required to recruit RITS to fission yeast centromeres and for maintenance of centromeric heterochromatin
- PMID: 18212052
- PMCID: PMC2268443
- DOI: 10.1128/MCB.01637-07
Chp1-Tas3 interaction is required to recruit RITS to fission yeast centromeres and for maintenance of centromeric heterochromatin
Abstract
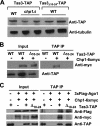
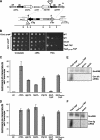
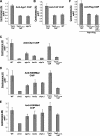
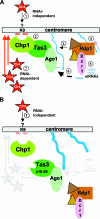
The maintenance of centromeric heterochromatin in fission yeast relies on the RNA interference-dependent complexes RITS (RNA-induced transcriptional silencing complex) and RDRC (RNA-directed RNA polymerase complex), which cooperate in a positive feedback loop to recruit high levels of histone H3 K9 methyltransferase activity to centromeres and to promote the assembly and maintenance of centromeric heterochromatin. However, it is unclear how these complexes are targeted to chromatin. RITS comprises Chp1, which binds K9-methylated histone H3; Ago1, which binds short interfering (siRNAs); the adaptor protein Tas3, which links Ago1 to Chp1; and centromeric siRNAs. We have generated mutants in RITS to determine the contribution of the two potential chromatin-targeting proteins Chp1 and Ago1 to the centromeric recruitment of RITS. Mutations in Tas3 that disrupt Ago1 binding are permissive for RITS recruitment and maintain centromeric heterochromatin, but the role of Tas3's interaction with Chp1 is unknown. Here, we define the Chp1 interaction domain of Tas3. A strain expressing a tas3 mutant that cannot bind Chp1 (Tas3(Delta)(10-24)) failed to maintain centromeric heterochromatin, with a loss of centromeric siRNAs, a failure to recruit RITS and RDRC to centromeres, and high levels of chromosome loss. These findings suggest a pivotal role for Chp1 and its association with Tas3 for the recruitment of RITS, RDRC, and histone H3 K9 methyltransferase activity to centromeres.
Figures

Similar articles
-
RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast.Mol Cell Biol. 2005 Mar;25(6):2331-46. doi: 10.1128/MCB.25.6.2331-2346.2005. Mol Cell Biol. 2005. PMID: 15743828 Free PMC article.
-
Continuous requirement for the Clr4 complex but not RNAi for centromeric heterochromatin assembly in fission yeast harboring a disrupted RITS complex.PLoS Genet. 2010 Oct 28;6(10):e1001174. doi: 10.1371/journal.pgen.1001174. PLoS Genet. 2010. PMID: 21060862 Free PMC article.
-
Studies on the mechanism of RNAi-dependent heterochromatin assembly.Cold Spring Harb Symp Quant Biol. 2006;71:461-71. doi: 10.1101/sqb.2006.71.044. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381328 Review.
-
An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing.Mol Cell. 2009 Apr 24;34(2):155-67. doi: 10.1016/j.molcel.2009.02.032. Mol Cell. 2009. PMID: 19394293 Free PMC article.
-
RITS-connecting transcription, RNA interference, and heterochromatin assembly in fission yeast.Wiley Interdiscip Rev RNA. 2011 Sep-Oct;2(5):632-46. doi: 10.1002/wrna.80. Epub 2011 Mar 23. Wiley Interdiscip Rev RNA. 2011. PMID: 21823226 Free PMC article. Review.
Cited by
-
High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin.Mol Cell. 2009 Apr 10;34(1):36-46. doi: 10.1016/j.molcel.2009.02.024. Mol Cell. 2009. PMID: 19362535 Free PMC article.
-
Chromosome segregation and organization are targets of 5'-Fluorouracil in eukaryotic cells.Cell Cycle. 2015;14(2):206-18. doi: 10.4161/15384101.2014.974425. Cell Cycle. 2015. PMID: 25483073 Free PMC article.
-
Control of Chromatin Structure by Long Noncoding RNA.Trends Cell Biol. 2015 Oct;25(10):623-632. doi: 10.1016/j.tcb.2015.07.002. Trends Cell Biol. 2015. PMID: 26410408 Free PMC article. Review.
-
Asterix/Gtsf1 links tRNAs and piRNA silencing of retrotransposons.Cell Rep. 2021 Mar 30;34(13):108914. doi: 10.1016/j.celrep.2021.108914. Cell Rep. 2021. PMID: 33789107 Free PMC article.
-
The Chp1 chromodomain binds the H3K9me tail and the nucleosome core to assemble heterochromatin.Cell Discov. 2016 Apr 19;2:16004. doi: 10.1038/celldisc.2016.4. eCollection 2016. Cell Discov. 2016. PMID: 27462451 Free PMC article.
References
-
- Allshire, R. C., J. P. Javerzat, N. J. Redhead, and G. Cranston. 1994. Position effect variegation at fission yeast centromeres. Cell 76157-169. - PubMed
-
- Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9218-233. - PubMed
-
- Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14943-951. - PubMed
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410120-124. - PubMed
-
- Bernard, P., and R. Allshire. 2002. Centromeres become unstuck without heterochromatin. Trends Cell Biol. 12419-424. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
