Expression of histone-U1 snRNA chimeric genes: U1 promoters are compatible with histone 3' end formation
- PMID: 1820206
- PMCID: PMC5952198
Expression of histone-U1 snRNA chimeric genes: U1 promoters are compatible with histone 3' end formation
Abstract
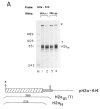
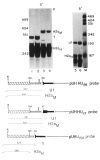
Chimeric genes which fuse the mouse histone H2a gene and the mouse U1b gene were constructed and introduced into CHO cells by cotransfection. In the UH genes, the U1b gene promoter and the start of the U1b gene were fused to the H2a gene in the 5' untranslated region. In the HU genes, the U1b 3' end was inserted into the 3' untranslated region of the H2a gene replacing the normal histone 3' end. Transcripts from the UH genes initiated at the start of the U1 gene and ended at the normal histone 3' end. Transcripts from the HU chimeric genes did not end at the U1 3' end but extended at least 80 nucleotides further and had heterogeneous 3' ends. Placing both a U1 snRNA promoter and a U1 snRNA 3' end around a histone coding region resulted in transcripts which initiate and terminate at the appropriate U1 ends. These results are consistent with previous reports that formation of the U1 3' ends require U1 promoters, but indicate that the histone 3' end can be formed on transcripts initiating at U1 promoters. The transcripts initiated at the U1 start site and ending at the histone 3' end are present on polyribosomes and show proper posttranscriptional regulation.
Figures

 AvaI;
AvaI;  NarI.
NarI.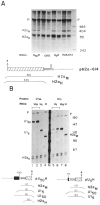




Similar articles
-
Efficient expression of protein coding genes from the murine U1 small nuclear RNA promoters.Proc Natl Acad Sci U S A. 1996 Aug 20;93(17):8852-7. doi: 10.1073/pnas.93.17.8852. Proc Natl Acad Sci U S A. 1996. PMID: 8799116 Free PMC article.
-
Increasing the distance between the snRNA promoter and the 3' box decreases the efficiency of snRNA 3'-end formation.Nucleic Acids Res. 1996 Nov 15;24(22):4525-34. doi: 10.1093/nar/24.22.4525. Nucleic Acids Res. 1996. PMID: 8948645 Free PMC article.
-
Formation of the 3' end of sea urchin U1 small nuclear RNA occurs independently of the conserved 3' box and on transcripts initiated from a histone promoter.Mol Cell Biol. 1992 Sep;12(9):4132-41. doi: 10.1128/mcb.12.9.4132-4141.1992. Mol Cell Biol. 1992. PMID: 1508209 Free PMC article.
-
Expression of human snRNA genes from beginning to end.Biochem Soc Trans. 2008 Aug;36(Pt 4):590-4. doi: 10.1042/BST0360590. Biochem Soc Trans. 2008. PMID: 18631122 Review.
-
The regulation of histone gene expression during the cell cycle.Biochim Biophys Acta. 1991 Mar 26;1088(3):327-39. doi: 10.1016/0167-4781(91)90122-3. Biochim Biophys Acta. 1991. PMID: 2015297 Review. No abstract available.
Cited by
-
Efficient expression of protein coding genes from the murine U1 small nuclear RNA promoters.Proc Natl Acad Sci U S A. 1996 Aug 20;93(17):8852-7. doi: 10.1073/pnas.93.17.8852. Proc Natl Acad Sci U S A. 1996. PMID: 8799116 Free PMC article.
-
Increasing the distance between the snRNA promoter and the 3' box decreases the efficiency of snRNA 3'-end formation.Nucleic Acids Res. 1996 Nov 15;24(22):4525-34. doi: 10.1093/nar/24.22.4525. Nucleic Acids Res. 1996. PMID: 8948645 Free PMC article.
-
Formation of the 3' end of sea urchin U1 small nuclear RNA occurs independently of the conserved 3' box and on transcripts initiated from a histone promoter.Mol Cell Biol. 1992 Sep;12(9):4132-41. doi: 10.1128/mcb.12.9.4132-4141.1992. Mol Cell Biol. 1992. PMID: 1508209 Free PMC article.
-
Point mutations in the stem-loop at the 3' end of mouse histone mRNA reduce expression by reducing the efficiency of 3' end formation.Mol Cell Biol. 1994 Mar;14(3):1709-20. doi: 10.1128/mcb.14.3.1709-1720.1994. Mol Cell Biol. 1994. PMID: 8114706 Free PMC article.
-
The histone mRNA 3' end is required for localization of histone mRNA to polyribosomes.Nucleic Acids Res. 1992 Nov 25;20(22):6057-66. doi: 10.1093/nar/20.22.6057. Nucleic Acids Res. 1992. PMID: 1461736 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
