Molecular evolution of the cytochrome c oxidase subunit 5A gene in primates
- PMID: 18197981
- PMCID: PMC2241769
- DOI: 10.1186/1471-2148-8-8
Molecular evolution of the cytochrome c oxidase subunit 5A gene in primates
Abstract
Background: Many electron transport chain (ETC) genes show accelerated rates of nonsynonymous nucleotide substitutions in anthropoid primate lineages, yet in non-anthropoid lineages the ETC proteins are typically highly conserved. Here, we test the hypothesis that COX5A, the ETC gene that encodes cytochrome c oxidase subunit 5A, shows a pattern of anthropoid-specific adaptive evolution, and investigate the distribution of this protein in catarrhine brains.
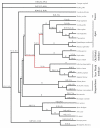
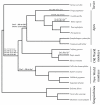
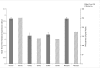
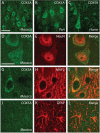
Results: In a dataset comprising 29 vertebrate taxa, including representatives from all major groups of primates, there is nearly 100% conservation of the COX5A amino acid sequence among extant, non-anthropoid placental mammals. The most recent common ancestor of these species lived about 100 million years (MY) ago. In contrast, anthropoid primates show markedly elevated rates of nonsynonymous evolution. In particular, branch site tests identify five positively selected codons in anthropoids, and ancestral reconstructions infer that substitutions in these codons occurred predominantly on stem lineages (anthropoid, ape and New World monkey) and on the human terminal branch. Examination of catarrhine brain samples by immunohistochemistry characterizes for the first time COX5A protein distribution in the primate neocortex, and suggests that the protein is most abundant in the mitochondria of large-size projection neurons. Real time quantitative PCR supports previous microarray results showing COX5A is expressed in cerebral cortical tissue at a higher level in human than in chimpanzee or gorilla.
Conclusion: Taken together, these results suggest that both protein structural and gene regulatory changes contributed to COX5A evolution during humankind's ancestry. Furthermore, these findings are consistent with the hypothesis that adaptations in ETC genes contributed to the emergence of the energetically expensive anthropoid neocortex.
Figures
Similar articles
-
Molecular evolution of cytochrome c oxidase subunit I in primates: is there coevolution between mitochondrial and nuclear genomes?Mol Phylogenet Evol. 2000 Nov;17(2):294-304. doi: 10.1006/mpev.2000.0833. Mol Phylogenet Evol. 2000. PMID: 11083942
-
Molecular evolution of aerobic energy metabolism in primates.Mol Phylogenet Evol. 2001 Jan;18(1):26-36. doi: 10.1006/mpev.2000.0890. Mol Phylogenet Evol. 2001. PMID: 11161739 Review.
-
Molecular evolution of cytochrome c oxidase subunit IV: evidence for positive selection in simian primates.J Mol Evol. 1997 May;44(5):477-91. doi: 10.1007/pl00006172. J Mol Evol. 1997. PMID: 9115172
-
Adaptive evolution of cytochrome c oxidase subunit VIII in anthropoid primates.Proc Natl Acad Sci U S A. 2003 May 13;100(10):5873-8. doi: 10.1073/pnas.0931463100. Epub 2003 Apr 25. Proc Natl Acad Sci U S A. 2003. PMID: 12716970 Free PMC article.
-
The evolution of brains from early mammals to humans.Wiley Interdiscip Rev Cogn Sci. 2013 Jan;4(1):33-45. doi: 10.1002/wcs.1206. Epub 2012 Nov 8. Wiley Interdiscip Rev Cogn Sci. 2013. PMID: 23529256 Free PMC article. Review.
Cited by
-
Studies of aging nonhuman primates illuminate the etiology of early-stage Alzheimer's-like neuropathology: An evolutionary perspective.Am J Primatol. 2021 Nov;83(11):e23254. doi: 10.1002/ajp.23254. Epub 2021 May 7. Am J Primatol. 2021. PMID: 33960505 Free PMC article. Review.
-
Comparative physiological anthropogeny: exploring molecular underpinnings of distinctly human phenotypes.Physiol Rev. 2023 Jul 1;103(3):2171-2229. doi: 10.1152/physrev.00040.2021. Epub 2023 Jan 5. Physiol Rev. 2023. PMID: 36603157 Free PMC article. Review.
-
A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing.Cell. 2008 Aug 8;134(3):416-26. doi: 10.1016/j.cell.2008.06.021. Cell. 2008. PMID: 18692465 Free PMC article.
-
The human brain: rewired and running hot.Ann N Y Acad Sci. 2011 May;1225 Suppl 1(Suppl 1):E182-91. doi: 10.1111/j.1749-6632.2011.06001.x. Ann N Y Acad Sci. 2011. PMID: 21599696 Free PMC article.
-
High-throughput RNA sequencing reveals structural differences of orthologous brain-expressed genes between western lowland gorillas and humans.J Comp Neurol. 2016 Feb 1;524(2):288-308. doi: 10.1002/cne.23843. Epub 2015 Aug 20. J Comp Neurol. 2016. PMID: 26132897 Free PMC article.
References
-
- Adkins RM, Honeycutt RL, Disotell TR. Evolution of eutherian cytochrome c oxidase subunit II: heterogeneous rates of protein evolution and altered interaction with cytochrome c. Molecular biology and evolution. 1996;13(10):1393–1404. - PubMed
-
- Andrews TD, Easteal S. Evolutionary rate acceleration of cytochrome c oxidase subunit I in simian primates. J Mol Evol. 2000;50(6):562–568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

