macroH2A1-dependent silencing of endogenous murine leukemia viruses
- PMID: 18195046
- PMCID: PMC2268408
- DOI: 10.1128/MCB.01362-07
macroH2A1-dependent silencing of endogenous murine leukemia viruses
Abstract
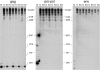
We show that macroH2A1 histone variants are important for repressing the expression of endogenous murine leukemia viruses (MLVs) in mouse liver. Intact MLV proviruses and proviruses with deletions in env were nearly silent in normal mouse liver and showed substantial derepression in macroH2A1 knockout liver. In contrast, MLV proviruses with a deletion in the 5' end of pro-pol were expressed in normal liver and showed relatively low levels of derepression in knockout liver. macroH2A1 nucleosomes were enriched on endogenous MLVs, with the highest enrichment occurring on the 5' end of pro-pol. The absence of macroH2A1 also led to a localized loss of DNA methylation on the 5' ends of MLV proviruses. These results demonstrate that macroH2A1 histones have a significant role in silencing endogenous MLVs in vivo and suggest that specific internal MLV sequences are targeted by a macroH2A1-dependent silencing mechanism.
Figures
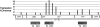


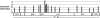
Similar articles
-
Antigenic subclasses of polytropic murine leukemia virus (MLV) isolates reflect three distinct groups of endogenous polytropic MLV-related sequences in NFS/N mice.J Virol. 2003 Oct;77(19):10327-38. doi: 10.1128/jvi.77.19.10327-10338.2003. J Virol. 2003. PMID: 12970417 Free PMC article.
-
Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation.Mol Cell Biol. 2001 Dec;21(23):7913-22. doi: 10.1128/MCB.21.23.7913-7922.2001. Mol Cell Biol. 2001. PMID: 11689684 Free PMC article.
-
Precise identification of endogenous proviruses of NFS/N mice participating in recombination with moloney ecotropic murine leukemia virus (MuLV) to generate polytropic MuLVs.J Virol. 2005 Apr;79(8):4664-71. doi: 10.1128/JVI.79.8.4664-4671.2005. J Virol. 2005. PMID: 15795252 Free PMC article.
-
Tag, you're hit: retroviral insertions identify genes involved in cancer.Trends Mol Med. 2003 Feb;9(2):43-5. doi: 10.1016/s1471-4914(03)00003-0. Trends Mol Med. 2003. PMID: 12615036 Review.
-
Silencing of Unintegrated Retroviral DNAs.Viruses. 2021 Nov 9;13(11):2248. doi: 10.3390/v13112248. Viruses. 2021. PMID: 34835055 Free PMC article. Review.
Cited by
-
Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues.Aging (Albany NY). 2013 Dec;5(12):867-83. doi: 10.18632/aging.100621. Aging (Albany NY). 2013. PMID: 24323947 Free PMC article.
-
The histone variant MacroH2A1 regulates target gene expression in part by recruiting the transcriptional coregulator PELP1.Mol Cell Biol. 2014 Jul;34(13):2437-49. doi: 10.1128/MCB.01315-13. Epub 2014 Apr 21. Mol Cell Biol. 2014. PMID: 24752897 Free PMC article.
-
Distinct distribution of ectopically expressed histone variants H2A.Bbd and MacroH2A in open and closed chromatin domains.PLoS One. 2012;7(10):e47157. doi: 10.1371/journal.pone.0047157. Epub 2012 Oct 30. PLoS One. 2012. PMID: 23118866 Free PMC article.
-
Macro domains as metabolite sensors on chromatin.Cell Mol Life Sci. 2013 May;70(9):1509-24. doi: 10.1007/s00018-013-1294-4. Epub 2013 Mar 3. Cell Mol Life Sci. 2013. PMID: 23455074 Free PMC article. Review.
-
Histone variants: dynamic punctuation in transcription.Genes Dev. 2014 Apr 1;28(7):672-82. doi: 10.1101/gad.238873.114. Genes Dev. 2014. PMID: 24696452 Free PMC article. Review.
References
-
- Abbott, D. W., M. Laszczak, J. D. Lewis, H. Su, S. C. Moore, M. Hills, S. Dimitrov, and J. Ausio. 2004. Structural characterization of macroH2A containing chromatin. Biochemistry 431352-1359. - PubMed
-
- Allen, M. D., A. M. Buckle, S. C. Cordell, J. Lowe, and M. Bycroft. 2003. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330503-511. - PubMed
-
- Aziz, D. C., Z. Hanna, and P. Jolicoeur. 1989. Severe immunodeficiency disease induced by a defective murine leukaemia virus. Nature 338505-508. - PubMed
-
- Barklis, E., R. C. Mulligan, and R. Jaenisch. 1986. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell 47391-399. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
